Introduction
The high-oleic trait is an important component of edible seed quality. Whether peanut is used as whole nuts, in peanut butter, or as a source of oil, fatty acid composition of the oil contributes to the stability of flavor and to health of the consumer. Due to lack of oxidative stability of double bonds, linoleic and linolenic acids oxidize more rapidly than does oleic acid, and this oxidation causes unpleasant odors and tastes (Bruner et al. 2001), limiting the storage quality of the peanut. Saturated fatty acids are stable to oxidation, but are not desired for health reasons (Wheeler et al., 1994; Bruner et al., 2001). Cis-unsaturated fatty acids (oleic, linoleic, and linolenic acids) have been claimed to be desirable for their role in lowering plasma cholesterol levels (Moore and Knauft, 1989). More recently, oleic acid has been shown to have desirable effects for health, reducing serum cholesterol levels and low density lipoprotein (LDL) levels (Kris-Etherton et al. 1999; O'Byrne et al. 1997; 1998).
In the past decade, several high-oleic peanut cultivars have been released. SunOleic 95R, developed by the University of Florida, was the first high-oleic peanut cultivar to be released (Gorbet and Knauft, 1997). It originated from a cross with a University of Florida high-oleic breeding line (UF435) and a component line of Sunrunner. The high-oleic runner FlavorRunner 458 has been the first high-oleic cultivar grown on a large scale, grown in West Texas but not elsewhere because of lack of resistance to disease and pests, especially resistance to tomato spotted wilt virus (TSWV), sclerotinia blight (Sclerotinia minor Jagger), and nematodes.
Several high-oleic varieties possessing various degrees of resistance to TSWV and Sclerotinia blight have been released recently. Among these are Tamrun OL01 (Simpson et al., 2003a) and Tamrun OL02, with moderate tolerance to TSWV and Sclerotinia blight. Tamrun OL01 is cultivated in parts of the southwestern United States and Oklahoma. Several newer releases include Andru II, GP-1, Norden, Hull, DP-1, and Georgia-02C (Branch, 2003). These all incorporate some degree of tolerance to TSWV, and some also have resistance to other diseases. The high-oleic Spanish variety ‘OLin’ incorporates resistance to Sclerotinia blight. However, no high-oleic variety incorporates resistance to nematodes.
Root-knot nematodes are the most economically-significant group of plant parasitic nematodes reported worldwide (Hussey and Janssen, 2001), causing serious problems on many cultivated plants. Within the root-knot group, Meloidogyne species M. incognita (Kofoid & White) Chitwood, M. javanica (Treub) Chitwood, M. arenaria (Neal) Chitwood, and M. hapla Chitwood) (Rodríguez-Kábana, 1997; Hussey and Janssen, 2001) are responsible for yield losses in many crops. M. hapla is limited to temperate soils and cool soils in the tropics, M. arenaria is found in tropical, subtropical and warm temperate soils, and M. javanica and M. incognita are typically tropical and subtropical (Whitehead, 1998). M. arenaria is economically the most important infesting parasite of peanut in the U.S. (Starr et al., 1990) although M. hapla, M. javanica, and M. haplanaria Eisenback (Eisenback et al., 2003) can also infect peanut (Abdel-Momen et al., 1998). M. arenaria infestation occurred in about 30% of the fields in Texas (Nelson et al., 1990), and the resulting yield losses were one of the major reasons for the shift of peanut hectarage away from this region.
Peanut producers have relied on crop rotation and nematicides for root-knot nematode control in peanuts, but host resistance, which suppresses nematode population of susceptible genotypes is probably the most economically-effective and environmentally-beneficial method of combating diseases and pests (Abdel-Momen et al., 1998; Russell, 1978). Nematode control is essentially prevention because once a plant is parasitized it is impossible to kill the nematode without destroying the host (Dufour et al., 1998). Nematode-resistant cultivars are developed by selection of plants with low nematode populations, which enables the plants to produce desirable yields in the presence of nematodes (Starr et al., 2002a).
Two nematode-resistant varieties ‘COAN’ and ‘NemaTAM’ have been released to date (Simpson and Starr, 2001; Simpson et al., 2003). Nematode resistance was transferred from A. cardenasii through a synthetic amphidiploid bridge (Simpson, 1991; Simpson et al., 1993). These runner varieties have a single dominant gene conferring resistance to M. arenaria and M. javanica, and yields are substantially higher than in infested fields than other varieties (Starr et al., 2002b.) RAPD (Random Amplification of Polymorphic Markers) and RFLP (Restriction Fragment Length Polymorphism) markers linked to this trait have been identified (Burow et al., 1996; Garcia et al., 1996; Choi et al. 1999) and were used for selection in the latter stages of development of NemaTAM (Simpson et al., 2003b).
Combining resistance to root-knot nematodes with high-oleic fatty acid content would allow more-profitable cultivation of high-oleic varieties in areas where nematode infestation is a serious impediment to peanut cultivation. In this paper, we present data from crosses resulting in progeny combining both traits, and on the genetics of these traits.
Materials and Methods
Combining high-oleic and root-knot nematode resistance traits .
To combine the high oleic/linoleic (O/L) ratio and nematode resistance traits, the high-oleic cultivars ‘OLin’ or ‘Tamrun OL01’ were hybridized with the cultivar ‘NemaTAM’, which is resistant to M. arenaria. A sample of 50 F2 seeds from each of the crosses was planted at the Texas Tech University Experimental Farm, Lubbock, TX in 2004 for collection of leaf samples for RFLP analysis.
Fatty Acid Analysis .
Fatty acid analysis of pieces of individual seeds of parents, F1 and F2 progeny was performed as per López et al. (2001). Oil analysis was performed using a Hewlett-Packard (HP) 5890 series II gas chromatograph with flame-ionization detector, model 7673 GC/SFC automatic injector and model 3396 series II integrator.
Fatty acid composition was measured on populations having a high-O/L and a low-O/L parent. For initial tests, seeds with an O/L ratio of 9∶1 or greater were considered to be in the high-oleic class, according to University of Florida patent #5,922,390 (Knauft et al., 1999.) All remaining seeds were classified as low-oleic. All parental seeds tested of high-oleic varieties had O/L ratios of >9∶1.
For further tests, the O/L ratios of F2 plants were classified as high-oleic, mid-oleic, or low-oleic by dividing the erstwhile low-O/L class into two groups according to parental values. A 95% confidence interval (CI) was calculated for the O/L ratio for the low-oleic parent NemaTAM. The F2 progeny having O/L ratio values less than the upper CI limit of the low-oleic parent were considered to be low oleic, and seeds with O/L values greater than 9∶1 were considered to be high-oleic. The remaining seeds were considered to be mid-oleic.
For testing F2 low∶high O/L segregation ratios, the χ2 statistic was used to test ratios of 3∶1 (low O/L∶high O/L, one gene), 15∶1 (low∶high, two genes), and 63∶1 (low∶high, three genes.) For testing segregation into 3 classes (low∶mid∶high O/L), 9∶6∶1 and 5∶10∶1 two-gene segregation ratios were tested in addition to the 15∶1 ratio. These additional ratios were used to test the hypotheses of epistatic and additive gene action. The 9∶6∶1 corresponded to epistasis, the ratio representing ‘dominance’ for both genes: only one gene with one or more ‘dominant’ alleles: all recessive alleles. The 5∶10∶1 ratio represented F2 progeny expected to possess three or four ‘dominant’ alleles: one or two ‘dominant’ alleles: all recessive alleles.
Determination of Nematode Resistance using RFLP Markers .
Three or four young unexpanded leaves from each F2 plant were collected, frozen in liquid nitrogen, and stored at −80 C. DNA extraction and Southern blotting were performed according to Burow et al. (2001). DNA was digested with EcoR I, and transferred to Hybond N+ membrane (Amersham, Inc) by alkalai transfer. Peanut probe R2545 (Choi et al., 1999; Burow et al., 2001) was generated from cDNA clones using the DIG High Prime DNA labeling and detection starter kit II (Roche Applied Science), according to the manufacturer's instructions, except as noted below. Insets amplified by polymerase chain reaction were precipitated using two volumes of ethanol, 1/10 volume of 5 M NaOH (pH 5.2), and 1μL of glycogen (5μg 5μL−1), and DNA was resuspended in 16–20∶L of TE. Between 300 ng and 1μg of template was used for probe synthesis. After incubation of probe and membranes overnight overnight at 42 C, membranes were washed twice with 100 mL of 2× SSC (saline-sodium citrate buffer) / 0.1% SDS (sodium dodecyl sulfate), once with 100 ml of 1× SSC/ 0.1% SDS, and once with 100 ml of 0.5× SSC/ 0.1% SDS; a washing temperature of 68 C was used for each wash of 15 min (Burow et al., 2001). A final wash was performed using 100 mL of washing buffer (0.1 M maleic acid, 0.15 M NaCl, pH 7.5, 0.3% (w/v) Tween 20), for five minutes at room temperature.
For development, membranes were transferred to clean Ziploc bags, blocked, treated with anti-digoxigenin-AP conjugate, washed, and developed with the CSPD reagent according to the manufacturer's instructions. The acetate sheet/ membrane/ acetate sheet sandwich was wrapped in Saran Wrap, incubated for 30 minutes at 37 C, and exposed to Kodak XAR X-ray film overnight before developing using a 1∶2 dilution of Kodak Dektol developer for 2 min, washing in indicator stop bath for 15 sec, and fixing using Kodak fixer for 2 min.
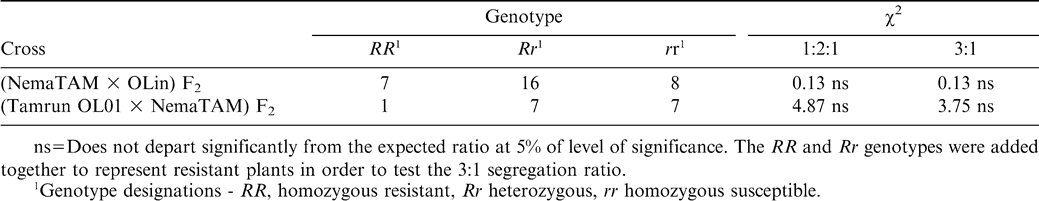
Plants having only the marker inherited from the dominant resistant parent were classified as homozygous resistant (RR); plants with one marker inherited from each parent were classified as heterozygous (Rr), and those plants with the marker inherited from susceptible parent were classified as homozygous recessive (rr), using comparison to the pattern published in Choi et al. (1999). Homozygous dominant and heterozygous genotypes were considered to be resistant, and the homozygous recessive genotypes were classified as susceptible. The χ2 test was used to test a 3∶1 (resistant∶susceptible) predicted F2 phenotypic segregation ratio, and a 1∶2∶1 genotypic segregation ratio was also tested.
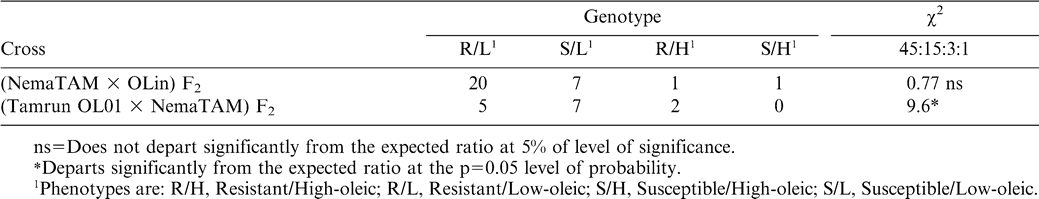
Cosegregation of nematode resistance and O/L ratio was tested against a predicted segregation ratio of 45∶15∶3∶1 (resistant/low-oleic∶susceptible/low-oleic∶resistant/high-oleic∶susceptible/high-oleic) for independently-assorting traits, assuming 3∶1 resistant∶susceptible and 15∶1 low∶high O/L ratios. The resistant (R) class was obtained by summing the RR and Rr marker genotypes; the susceptible (S) class was represented by the rr marker genotype.
Results
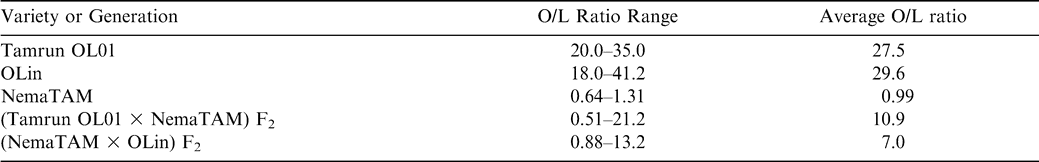
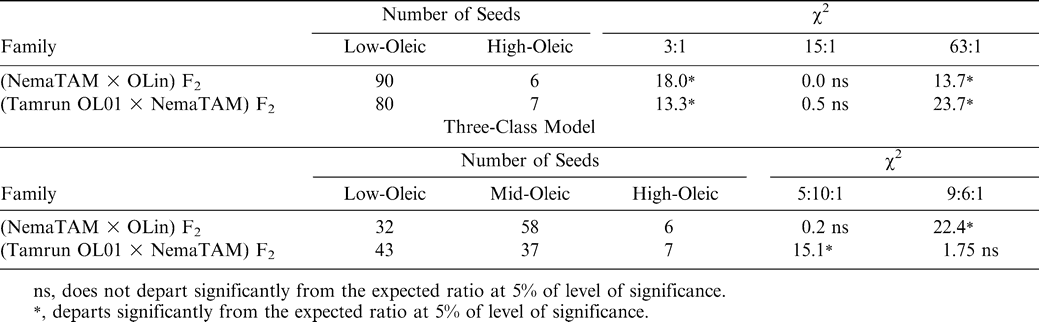
Analysis of F2 progeny of the Tamrun OL01 × NemaTAM and OLin × NemaTAM crosses demonstrated that the high-oleic trait was introduced into populations. The O/L ratios for the F2 progeny ranged from 0.51 to 21.2 in the crosses of Tamrun OL01 × NemaTAM, and 0.88 to 13.2 for the crosses of OLin × NemaTAM suggesting the reappearance of the high-oleic trait in the F2 generation (Table 1). Assuming that all progeny with F2 ratios < 9∶1 are low-oleic, the F2 progeny were consistent with a 2-gene recessive model for oil composition fitting the expected 15∶1 (low-oleic∶high-oleic) ratio as reported by Moore and Knauft (1989) (Table 2).
A more-careful examination of the data suggests that there is a class of mid-oleic progeny, and this class requires a different model of gene action than the two-gene recessive model. A statistical analysis of the low-oleic parent (NemaTAM) indicated that the 95% upper confidence interval for the O/L ratio of its seed was 1.79. Therefore seeds with low-O/L ratios similar to this parent should not exceed a 1.79∶1 ratio; seeds that fall between O/L ratios of 1.79 and 9.0 must belong to a mid-oleic class. Of 96 F2 seeds from the cross of NemaTAM × OLin, the O/L ratio of 58 fell into the mid-oleic range, as did 37 seeds of 87 from the cross of Tamrun OL01 × NemaTAM (Table 2; Figs. 1 and 2). A break between low-oleic and mid-oleic classes is evident in the NemaTam × OLin cross (Fig. 1); the break between classes is not pronounced in the Tamrun OL01 × NemaTAM cross (Fig. 2.)
The results of χ2 tests on the three-class model were consistent with different genetic models for the two crosses. An epistatic model of action was consistent with the data from the Tamrun OL01 × NemaTAM cross, fitting a two-gene 9∶6∶1 progeny ratio of low O/L∶mid O/L∶high O/L ratios. For the OLin × NemaTAM cross, the F2 progeny did not fit the 9∶6∶1 ratio, but did fit a 5∶10∶1 model. This latter is consistent with additive gene action, assuming this represents 3–4∶1–2∶0 alleles, respectively, for the low-oleic trait.
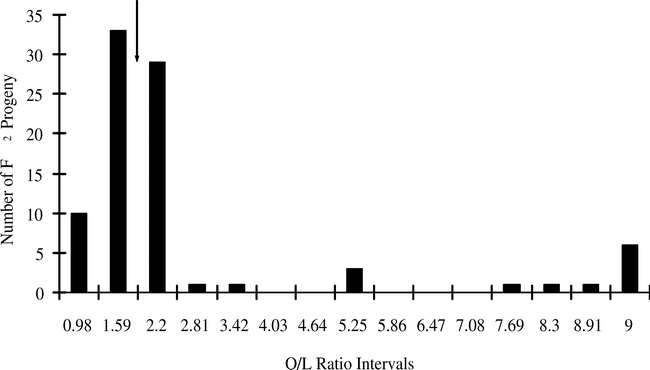
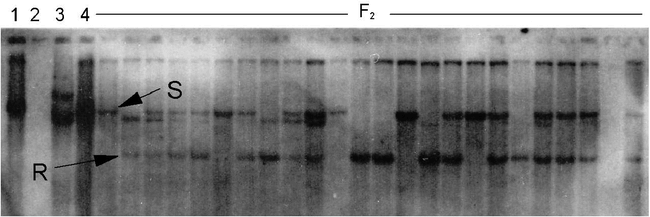
In addition to the high-O/L trait, nematode resistance was also introduced into both populations. Resistance was estimated using RFLP markers (Fig. 3), with resistant pattern being compared to Choi et al. (1999) as the source of the nematode resistance gene was the same as in the previous publications. In both crosses the predicted phenotypes fit a 3∶1 resistant∶susceptible model, in accordance with previous results that the root-knot nematode resistance trait is controlled by one dominant gene. Marker genotypes segregated in the predicted a 1∶2∶1 ratio of homozygous resistant∶heterozygous∶homozygous susceptible genotypes (Table 3). The homozygous resistant (RR) and heterozygous (Rr) classes were expected to be resistant, and rr was expected to be susceptible (Choi et. al., 1999).
RFLP marker pattern (Probe R2545) linked to the root-knot nematode resistance gene in peanuts.
The F2 genotypes are from the cross NemaTAM × OLin. Lanes denote: 1, Florunner; 2, Tamrun OL01 (insufficient DNA); 3, TxAG-6 (donor of the nematode resistance gene); 4, OLin; 5 to 28, F2 progeny. The letters ‘R’ and ‘S’ denote markers for the resistant and susceptible alleles, respectively, as determined by Choi et al. (1999).
Progeny combining nematode resistance and the high-oleic trait were present in F2 progeny of both crosses. A 3/64 proportion of the F2 progeny were expected to be high-oleic and resistant, three progeny of 43 tested were in this group. Co-segregation of F2 progeny for the high-oleic and nematode resistance traits were tested against a 45∶15∶3∶1 (resistant/ low-oleic∶ susceptible/ low-oleic∶ resistant/ high-oleic∶ susceptible/high-oleic) segregation ratio (Table 4). The NemaTAM × OLin cross fit the expected ratio, but the Tamrun OL01 × NemaTAM cross did not. However, the number of progeny tested from the latter cross was small; nevertheless it was in the two classes with the highest expected number of progeny that there was the largest deviation from expected, there being a deficiency in the resistant low-oleic class.
Discussion
Combination of high-oleic fatty acid composition and nematode resistance .
Through hybridization of high-oleic and nematode-resistant parents, these two traits have been combined in single F2 plants. Segregation of nematode resistance fit the single gene dominant model proposed previously, and the high-oleic trait appeared to require the presence of two recessive genes to be expressed. As such, 3/64 of the F2 progeny were expected to possess both traits if the traits were unlinked, and this proportion was borne out experimentally, as three of 43 plants were both resistant and high-oleic.
The relationship between the high-oleic and nematode resistance traits is unclear. In one cross (NemaTAM × OLin), there was no evidence for linkage, but in the other (Tamrun OL01 × NemaTAM), the traits did not segregate independently. Further work will be needed to clarify the issue. It is possible that the small number of progeny tested from this cross may be associated with chance deviation from the expected ratio. Alternatively, if the traits are linked, the difference between crosses may be associated with different genes for the high-oleic trait.
Additional resistant, high-oleic progeny can be expected in the F3 and successive generations if resistant, mid-oleic progeny are self-pollinated. This is because the models (see below) explaining this class involve heterozygosity at one or more loci controlling this trait.
Definition of the mid-oleic class and mode of gene action .
The high oleic/linoleic fatty acid trait in peanut has been reported to be controlled by two recessive genes (Moore and Knauft, 1989), with high-oleic and low-oleic classes. Data on the segregation of the trait in the crosses presented in the current work did not fit this two-class model. Statistical treatment of low-oleic and high-oleic parents and comparison to the F2 population demonstrated that significant proportions of F2 individuals fell into an intermediate range, referred to as mid-oleic. This is in accordance with López et al. (2001), who demonstrated the presence of a class of mid-oleic seeds. In the current experiment, one cross fit the 9∶6∶1 epistatic model and the other fit the 5∶10∶1 additive model. As López et al. (2001) postulated the presence of from 4 to 6 genes controlling the high-oleic trait, it is possible that differing mechanisms of gene action are present in the two crosses.
Data from other crop species have given evidence for both epistatic and additive mechanisms of inheritance of oil composition. In castor, inheritance of the high-oleic trait segregated in a 13∶3 fashion, consistent with control by two epistatic genes (Rojas-Barros et al., 2005). In Brassica, two epistatic quantitative trait loci (QTLs) were associated with erucic acid content (Mahmood et al., 2003). Eicosenoic acid content fell into high, mid, and low-eicosenoic groups, and these followed additive inheritance depending on the number of alleles for erucic acid. Three additional QTLs were associated with linoleic acid content. In B. napus, five additive alleles governing erucic acid content have been identified (Krzymanski and Downey, 1969).
Summary and Conclusions
Combination of the nematode resistance gene and high oleic trait along with desirable agronomic traits in a single variety will require a large initial population or multiple cycles of backcrossing and selection because most progeny will not possess both traits. However, recognition and use of resistant plants in the mid-oleic category can be beneficial to a breeding effort. Initial selection of this class can allow a larger population for selection of desirable agronomic traits, with the possibility of recovering high-oleic progeny in later generations by screening selfed progeny for this trait.
Acknowledgements
Graduate support for A. Muitia was provided by the award “Capacity Building in Public Sector Research and Extension” USAID/Mozambique, grant 656-G-00-00-00050-00, administered by the International Sorghum and Millet Cooperative. The authors would like to thank Mr. Jamie Ayers for assistance in maintenance of field plots.
Literature Cited
Abdel-Momen S. M. , Simpson C. E. , and Starr J. L. 1998 Resistance of interspecific Arachis breeding lines to Meloidogyne javanica and an undescribed Meloidogyne species. J. Nematol 30 : 341 – 346 .
Branch W. D. 2003 Registration of ‘Georgia-02C’ peanut. Crop Sci 43 : 1883 – 1884 .
Bruner A. C. , Jung S. , Abbot A. G. , and Powell G. L. 2001 The naturally occurring high oleate oil character in some peanut varieties results from reduced oleoyl-pc desaturase activity from mutation of aspartate 150 to asparagine. Crop Sci 41 : 522 – 526 .
Burow M. D. , Simpson C. E. , Paterson A. H. , and Starr J. L. 1996 Identification of peanut (Arachis hypogaea L.) RAPD markers diagnostic of root-knot nematode (Meloidogyne arenaria (Neal) Chitwood) resistance. Mol. Breeding 2 : 369 – 379 .
Burow M. D. , Simpson C. E. , Starr J. L. , and Paterson A. H. 2001 Transmission genetics of chromatin from a synthetic amphidiploid to cultivated peanut (Arachis hypogaea L.): broadening the gene pool of a monophyletic polyploid species. Genetics 159 : 823 – 837 .
Choi K. , Burow M. D. , Church G. , Burow G. , Paterson A. H. , Simpson C. E. , and Starr J. L. 1999 Genetics and mechanisms of resistance to Meloidogyne arenaria. In: Peanut Germplasm. J. Nematol 31 : 283 – 290 .
Dufour R. , Earles R. , Kuepper G. , and Greer L. 1998 Alternative Nematode Control. Fayetteville AR. URL:<http://attra.ncat.org/attra-pub/PDF/nematode.pdf〉. Accessed in August, 2004.
Eisenback J. D. , Bernard E. C. , Starr J. L. , Lee T. A. , and Tomaszewski E. K. 2003 Meloidogyne haplanaria n. sp (Nematodai meloidogynidae), a root-knot nematode parasitizing peanut in Texas. J. Nematol 35 : 395 – 403 .
Garcia G. M. , Stalker H. T. , Shroeder E. , and Kochert G. 1996 Identification of RAPD, SCAR, and RFLP markers tightly linked to nematode resistance genes introgressed from Arachis cardenansii into Arachis hypogaea. Genome 39 : 836 – 845 .
Gorbet D. W. and Knauft D. A. 1997 Registration of ‘SunOleic 95R’ peanut. Crop Sci 37 : 1392 .
Hussey R. S. and Janssen G. J. W. 2001 Root-knot nematodes: Meloidogyne species. 43 – 65 In: Starr J. L. , Cook R. , and Bridge J. eds. Plant resistance to parasitic nematodes CABI Publishing London .
Knauft D. A. , Gorbet D. W. , Norden A. J. , and Norden K. C. 1999 Peanut oil from enhanced peanut products United States Patent #5,922,390. Available at URL:<http://patft.uspto.gov/netahtml/srchnum.htm〉. Verified on November 27, 2004.
Kris-Etherton P. M. , Pearson T. A. , Wan Y. , Hargrove R. L. , Moriarty K. , Fishell V. , and Etherton T. D. 1999 High-monounsaturated fatty acid diets lower both plasma cholesterol and triacylglycerol concentrations. Am. J. Clin. Nutr 70 : 1009 – 15 .
Krzymanski J. and Downey R. K. 1969 Inheritance of fatty acid composition in winter forms of rapeseed, Brassica napus. Can. J. Plant Sci 49 : 313 – 319 .
López Y. , Smith O. D. , Senseman S. A. , and Rooney W. L. 2001 Genetic factors influencing high-oleic acid content in Spanish market-type peanut cultivars. Crop Sci 41 : 51 – 56 .
Mahmood T. , Ekuere U. , Yeh F. , Good A. G. , and Stringam G. R. 2003 RFLP linkage analysis and mapping genes controlling the fatty acid profile of Brassica juncea using reciprocal DH populations. Theor. Appl. Genet 107 : 283 – 290 .
Moore K. M. and Knauft D. A. 1989 The inheritance of high-oleic acid in peanut. J. Hered 80 : 252 – 253 .
Nelson S. C. , Simpson C. E. , and Starr J. L. 1990 Expression of resistance to Meloidogyne arenaria in Arachis batizicoi and A. cardenasii. J. Nematol 22 : 423 – 425 .
O'Byrne D. J. , Knauft D. A. , and Shireman R. B. 1997 Low fat-monounsaturated rich diets containing high-oleic peanuts improve serum lipoprotein profiles. Lipids 32 : 687 – 695 .
O'Byrne D. J. , O'Keefe S. F. , and Shireman R. B. 1998 Low-fat, monounsaturate-rich diets reduce susceptibility of low density lipoproteins to peroxidation ex vivo. Lipids 33 : 149 – 157 .
Rodríguez-Kábana R. 1997 Nematode diseases. 45 – 53 in Kokalis-Burelle N. , Porter D. M. , Rodríguez-Kábana R. , Smith D. H. , and Subrahmanyam P. eds. Compendium of peanut diseases, second edition The American Phytopathological Society St. Paul .
Rojas-Barros P. , de Haro A. , and Fernández-Martínez J. M. 2005 Inheritance of high oleic/ low ricinoleic acid content in the seed oil of castor mutant OLE-1. Crop Sci 45 : 157 – 162 .
Russell G. E. 1978 Plant breeding for pest and disease resistance Butterworth Co London 12 – 35 – 41 .
Simpson C. E. 1991 Pathways for introgression of pest resistance into Arachis hypogaea. Peanut Sci 18 : 22 – 26 .
Simpson C. E. , Baring M. R. , Schubert A. M. , Melouk H. A. , Black M. C. , Lopez Y. , and Keim K. A. 2003a Registration of ‘Tamrun OL01’ peanut. Crop Sci 43 : 2298 .
Simpson C. E. , Nelson S. C. , and Starr J. L. 1993 Registration of TxAG-6 and TxAG-7 peanut germplasm lines. Crop Sci 33 : 1418 .
Simpson C. E. and Starr J. L. 2001 Registration of ‘COAN’ peanut. Crop Sci 41 : 918 .
Simpson C. E. , Starr J. L. , Church G. L. , Burow M. D. , and Paterson A. H. 2003b Registration of ‘NemaTAM’ peanut. Crop Sci 43 : 1561 .
Starr J. L. , Schuster G. L. , and Simpson C. E. 1990 Characterization of resistance to Meloidogyne arenaria in an interspecific Arachis spp. hybrid. Peanut Sci 17 : 106 – 108 .
Starr J. L. , Morgan E. R. , and Simpson C. E. 2002a Management of the peanut root-knot nematode, Meloidogyne arenaria, with host resistance Plant Management Network St. Paul Available at URL:<http://www.plantmanagementnetwork.org/pub/php/ management/rootknot〉 Verified Sept. 14, 2005.
Starr J. L. , Bridge J. , and Cook R. 2002b Resistance to plant-parasitic nematodes: history, current use and future potential. In: Starr J. L. , Bridge J. , and Cook R. eds. Plant resistance to parasitic nematodes CABI Publishing London .
Wheeler R. A. , Smith R. L. , and Knauft D. A. 1994 Microsomal polypeptide comparisons between high and normal oleic acid isogenic peanut lines using two-dimensional gel electrophoresis. Peanut Sci 22 : 75 – 78 .
Whitehead A. G. 1998 Plant Nematode Control CAB International London .
Notes
- Texas Tech University, Department of Plant and Soil Science, Lubbock, TX 79409. [^]
- Texas Agricultural Experiment Station, Department of Soil and Crop Science, Texas AM University, Lubbock, TX 79403. [^]
- Texas AM University, Department of Plant Pathology Microbiology, College Station, TX 77843. [^]
- *Current Address: Lichinga Research Station, P.O. Box 238, Lichinga, Niassa, Mozambique. [^]
- **Work performed in partial fulfillment of the Master's of Science in Crop Science degree. [^] ***Corresponding author, address: Texas Agricultural Experiment Station, 1102 East FM 1294, Lubbock, TX 79403, Phone: (806)-746-6101, FAX: (806)-746-6528, Email: mburow@tamu.edu
Author Affiliations