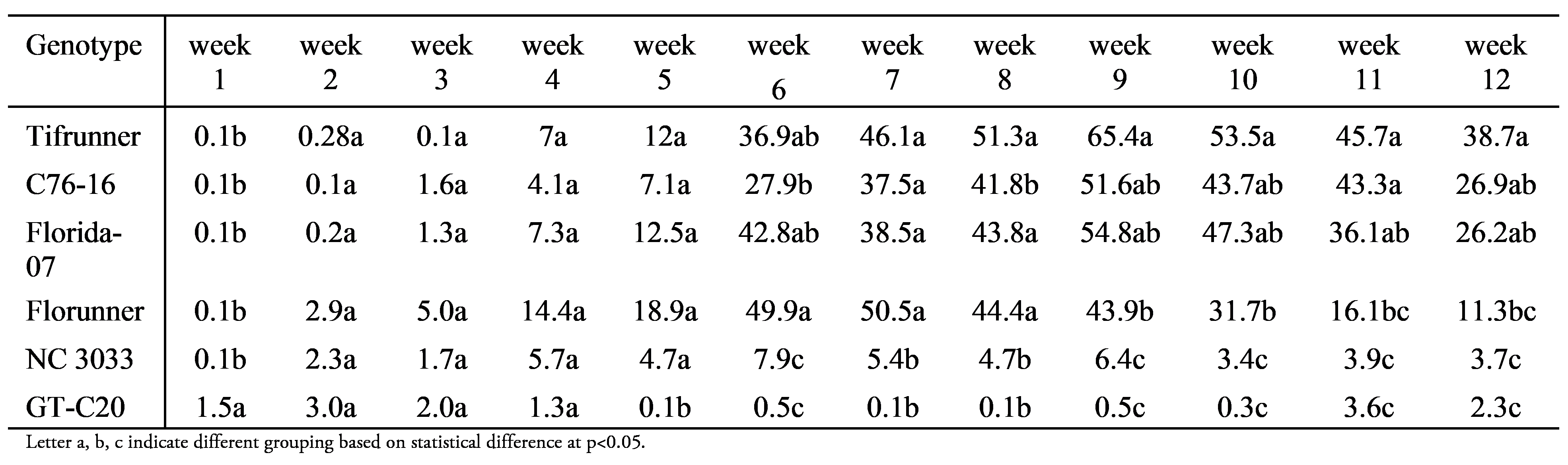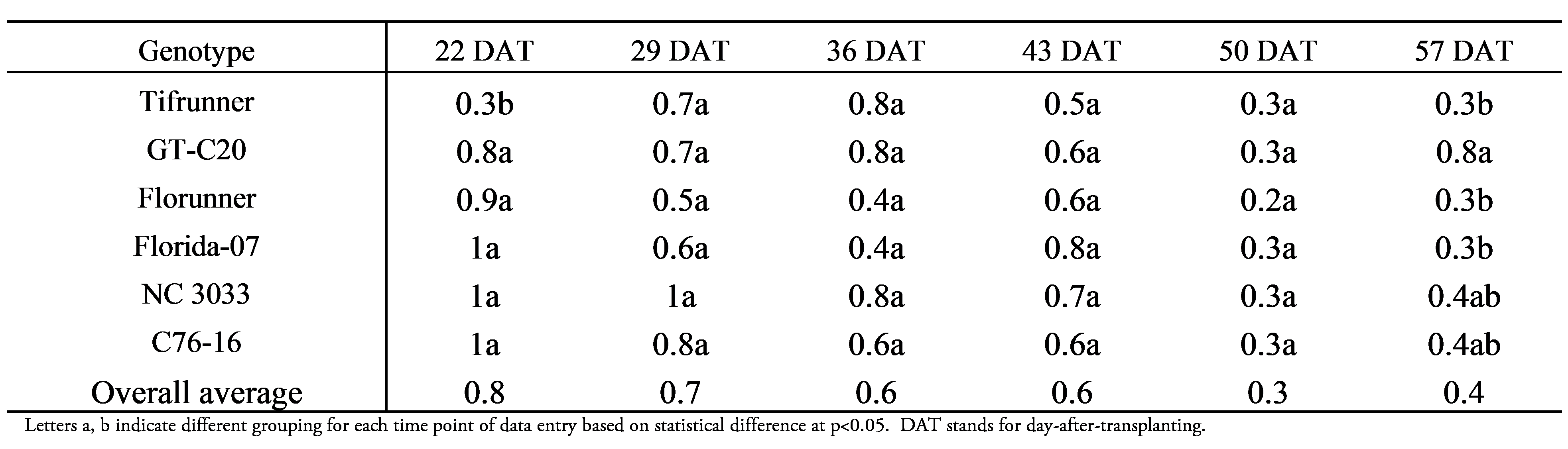INTRODUCTION
Cultivated peanut is widely grown in tropical and subtropical regions globally and 48 million tons of in-shell peanut were produced in 2020 (faostat.org). Highly diverse growing environments across the world demand that peanut cultivars are well adapted to local conditions. US peanut production is completely mechanized and requires significant inputs including irrigation and disease management (Holbrook and Stalker, 2002). Medium to late maturing peanut cultivars dominate the southeastern US growing regions due to their high yield potential associated with a protracted season of pod fill. On the other hand, peanut production in Africa and India occurs in areas with limited access to irrigation (Nigam and Aruna, 2008; Holbrook et al., 2016). Under the constraints of irregular and insufficient rainfall, short-duration cultivars are needed to escape drought stress. For regions in the US with short growing seasons, early maturing cultivars ensure harvest before the threat of frost. Therefore, peanut cultivars with time-to-maturity (the length of time required for peanut to reach maturity) ranging from 80 to 180 days after planting have been selected and grown worldwide in their adaptive range (Stalker and Simpson 1995).
Peanut has an indeterminate growth habit since its apical meristems never undergo a vegetative to floral transition. In fact, it has been shown that ‘Florunner’ peanut can exhibit a perennial type of growth when protected from disease and temperature stresses (Kvien and Ozias-Akins, 1991). Seeds that develop on a peanut plant display a range of maturity at harvest (Pattee et al., 1974). Plants with 60 to 80% of developed pods demonstrating brown to black inner pericarp coloration are generally considered mature (Williams and Drexler, 1981; Rowland et al., 2006). Underground development of peanut pods makes it difficult to evaluate fruit maturity before harvest. The hull scrape method was developed and implemented as a peanut profile board (Williams and Drexler, 1981) which has been widely used to determine optimal maturity for harvest. While defining time-to-maturity for a peanut cultivar, consideration must be given not only to maximize maturity but also economic returns (Sorensen et al., 2020). Harvesting too early often results in a high percentage of immature pods and subsequent high risk of post-harvest aflatoxin contamination (Rucker et al., 1994). In addition, low percentages of sound and mature kernels from early harvest decreases peanut price (Lamb et al., 2010). On the contrary, late harvest can result in yield loss due to pod loss from peg decay and/or seed germination.
Although time-to-maturity is critical to peanut adaptability, yield and quality, little is known regarding the genetic controls of this trait. One study indicated relative low heritability of this trait (Ali and Wynne, 1994) and few QTL associated with maturity index were reported from two recombinant inbred line (RIL) populations (Chirinos, 2011). Most recently, two reproducible QTL on chromosomes A04 and B03 explaining 9.3% and 11.9% of phenotypic variation for maturity index were identified in a RIL population created from two virginia-type peanuts (Kunta et al., 2021). The lack of sufficient genetic information on time-to-maturity partly is due to the complexity of genetic components contributing to the sum of phenotypic expression of time-to-maturity. It is known that flowering time and plant architecture contribute to time-to-maturity in legume species (Huyghe, 1998). Deconstructing the total phenotype of time-to-maturity into component traits might improve the resolution of phenotyping and provide data to explore multiple genetic pathways influencing time-to-maturity.
Based on morphological characteristics, such as the presence of flowers on the main stem and growth habit, cultivated peanut has been classified into two subspecies, hypogaea and fastigiata; and six botanical varieties, hypogaea, hirsuta, fastigiata, vulgaris, aequatoriana, and peruviana. In addition, this species is also divided into four market types: runner, virginia, spanish, and valencia (Krapovickas and Gregory, 2007; Stalker, 2017). Runner and virginia market types belong to subsp. hypogaea which produces no flowers on the main stem and exhibits alternating vegetative and reproductive nodes on the laterals. The growth habit of runner and virginia peanuts tends to be spreading or bunch. The main distinction between runner and virginia peanut lines is in the seed size in which the latter is larger than the former. Spanish and valencia market types are in the subsp. fastigiata that is characterized by the presence of flowers on the main stem and sequential reproductive nodes on the laterals. Most spanish and valencia peanuts have an erect or bunch type of growth habit. Therefore, it will be beneficial to take separate measurements of component traits including flowering on the main stem, flowering rate, internode length, branching patterns, flower-to-peg conversion ratio, percentage of harvestable pods and harvest index to capture the genetic factors contributing to the variation in time-to-maturity among cultivated peanut.
Materials and Methods
Genetic materials
In this study, six peanut genotypes were investigated, ‘Tifrunner’ (Holbrook and Culbreath, 2007), GT-C20 (Liang et al., 2006), Florunner (Norden et al., 1969), ‘Florida-07’ (Gorbet and Tillman, 2008), NC 3033 (Beute et al., 1976), and C76-16 (Holbrook et al., 2007). These genotypes were chosen because several were parents of seven RIL populations (Table S1) (Holbrook et al., 2013; Qin et al., 2012). The Tifrunner and GT-C20 reciprocal population was developed independently from the published T-population with the same parents (Hong et al., 2010; Wang et al., 2013). The remaining six populations were part of the peanut nested association mapping population (Holbrook et al., 2013; Chu et al., 2018).
Tifrunner is a tomato spotted wilt virus (TSWV) resistant cultivar with spreading runner growth habit and a prominent main stem. Tifrunner was the reference genotype for the first runner-type whole genome sequence (Bertioli et al., 2019). GT-C20 is a spanish peanut cultivar with a bunch growth habit and early maturity. Florunner has a spreading growth habit with typically alternate pairs of reproductive and vegetative nodes on the laterals. Florida-07 is a high-yielding runner-type peanut with strong resistance to multiple diseases. NC 3033 is germplasm derived from a virginia by spanish cross. C76-16 is a runner-type germplasm with improved resistance to drought and aflatoxin contamination. Phenotypic variation among the genotypes was determined for the component traits of time-to-maturity.
Flower-to-peg conversion and yield
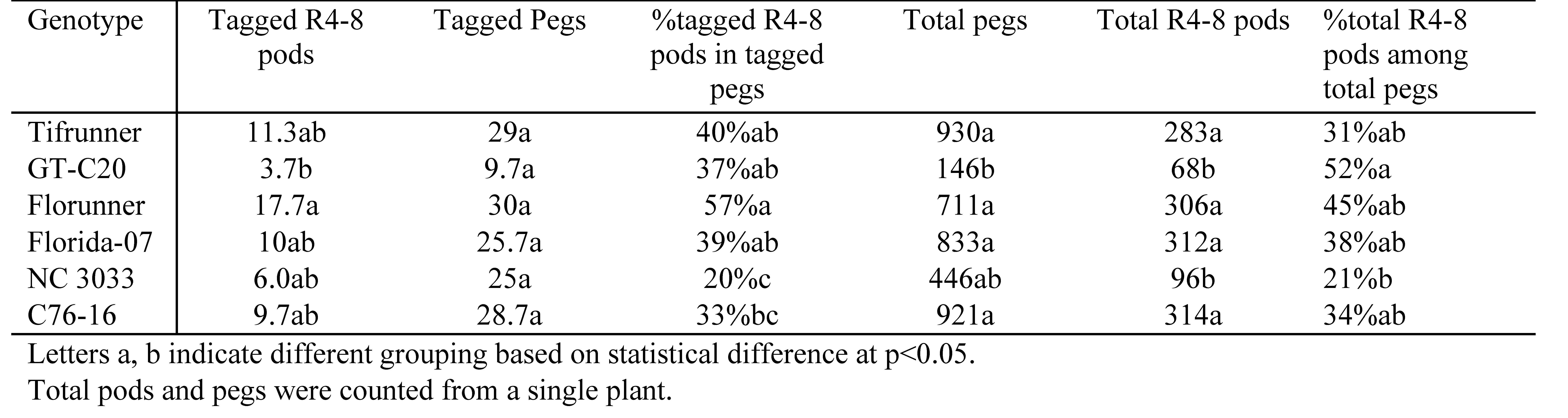
The six peanut genotypes were sown in Jiffy pots ( www.jiffypots.com) and germinated in the greenhouse on April 26, 2018. Eighteen days after planting, seedlings were transplanted to the field located on the UGA Tifton Campus, Tifton, Georgia (31.4746° N, 83.5308° W). Seedlings were planted in the center of two-meter-wide beds, one meter apart from each other. Distance of plants between neighboring beds was four meters. Eighteen randomized complete blocks were dedicated for data collection. Scouting for flowers was performed 10 days after transplanting (DAT) and tagging of flowers was initiated when more than 80% of the plants produced their first flower. Three blocks were used for tagging on each of the six dates, i.e., 22, 29, 36, 43, 50, and 57 DAT. Therefore, all plants in this experiment were tagged only once to minimize physical damage to tagged pegs. On each date, all flowers from the three blocks were tagged with cotton threads. Ten days later, pegs initiated from the tagged flowers were carefully threaded with the cotton threads. All of the tagged pegs and harvestable pods at R4-8 maturity stages were counted upon harvest. The percentage of tagged pegs over tagged flowers at each time point was calculated to represent flower-to-peg conversion rate. The percentage of tagged harvested pods over total number of tagged pegs and pods represent peg to pod conversion rate.
Right before harvest, two measurements of the canopy were taken; one measurement was taken for width of the canopy across the center of the canopy and the second measurement was taken at an angle perpendicular to the width and across the center of the canopy. The area of the canopy was estimated by the product of diagonal lengths of the plant divided by two which is the area formula for rhombus. Harvest dates were selected when more than 70% of the randomly sampled five to 10 pods showed coloration in the mesocarp of the saddle area of the pods (Williams and Drexler, 1981). Harvest dates for each genotype were as follows, GT-C20, 113 DAP; NC 3033, 125 DAP; Florunner, 132 DAP; C76-16, 133 DAP; Tifrunner and Florida-07, 134 DAP. Temperature and rainfall data were downloaded from the Tifton site of www.georgiaweather.net) (Figure S1). Six time points of flower tagging and harvest dates of all genotypes were marked in the chart. Cumulative GDD was determined by Σ(Tavg – Tbase) where Tavg is the daily average temperature and Tbase is 13 C, below this temperature peanut growth is arrested (Oakes et al., 2020).
Upon harvest, above ground tissue of each plant was collected. All pods and pegs were separated from the canopy. Pegs at maturity stages R2-R3 and pods at R4-R8 stages (Boote, 1982) were counted separately. R4-R8 stage pods were considered as harvestable pods since these pods are fully expanded. The total number of pegs per plant was calculated by the sum of these two sets of counts. R4-R8 stage pods and canopy were air dried for two weeks before collecting the weight data. Harvest index (HI) was calculated by dividing dried pod weight by dried pod plus canopy weight. R2-R3 stage pegs and R4-R8 stage pods with thread tags were documented as well. Fifty R4-R8 pods were weighed and hand-shelled. Mature pods were identified by the presence of dark blotches on the inner pericarp of pods (Gilman and Smith, 1977). Seeds from mature pods were counted as mature seeds. Weight of both mature and immature pods and seeds were taken. Total pod and seed number from each plant were counted separately. Percent of mature pod number was calculated by dividing the number of mature pods by the total pod number. Percent of mature seed number was calculated by dividing the number of mature seeds by the total seed number. Percent of mature pod weight was calculated by dividing the weight of mature pods by the total pod weight. Percent of mature seed weight was calculated by dividing the weight of mature seeds by the total seed weight.
The axil of each node on the two cotyledonary primary laterals (n+1_c laterals) was inspected from the basal end joining the main stem to the distal terminal. A reproductive node was documented when dried flowers and/or pegs were present at the axil. A node was documented as vegetative when there was an absence of flowers/pegs at the axil. The percentage of reproductive nodes over the total of reproductive and vegetative nodes was calculated. In addition, alternating points between reproductive and vegetative nodes were counted using the “if” function of Excel software. Alternating frequency was calculated as the ratio of alternating points over the sum of reproductive and vegetative nodes. Low alternating frequency suggests that nodes of the same type form a consecutive distribution pattern. High alternating frequency indicates that the two types of nodes tend to alternate at a high rate. Additionally, length and total node number of n+1_c laterals were documented. The internode length of n+1_c laterals was calculated by dividing the branch length by node number. Branch numbers on n+1, n+2, and n+3 laterals also were documented.
Number of flowers throughout the growing season
In order to document the number of flowers produced throughout the growing season, a separate set of seedlings of the above six genotypes was transplanted in the same manner. Seven field replicates were included following a randomized complete block design. Flowers were counted three times a week for 12 weeks starting from 11 DAT. Weeks two to seven overlapped with the period for the flower tagging experiment described above. Total flower number and distribution of flowers on branches affect reproductive efficiency and time-to-maturity in peanuts (Coffelt et al., 1989; Giayetto et al., 2013). Flowers formed early and at positions close to the soil have a high probability to produce mature pods. To account for the branching patterns in peanut, the main stem is conventionally noted as n and all branches arising from the nodes on the main stem are noted as n+1. Any branches arising from nodes on the n+1 branch were called n+2 and so forth (Shashidhar et al., 1986). The two n+1 lateral branches that developed at each axil of the cotyledonary node are called cotyledonary laterals (n+1_c). It has been shown that pods formed on cotyledonary laterals accounted for 70 to 90% of pod production in spanish type peanuts (Choudhari et al., 1985; Shashidhar et al., 1986). Therefore, flower numbers on the main stem (n), n+1_c, and other laterals were counted separately in this study.
ANOVA analysis was performed for data sets collected at harvest. Data sets collected as a time series were analyzed by repeated measure using the PROC GLIMMIX procedure. Statistical analysis was performed with SAS ® Enterprise Guide 7. Data entry was performed with FieldBook, an open-source application (Rife and Poland, 2014).
Results and discussion
Number of flowers
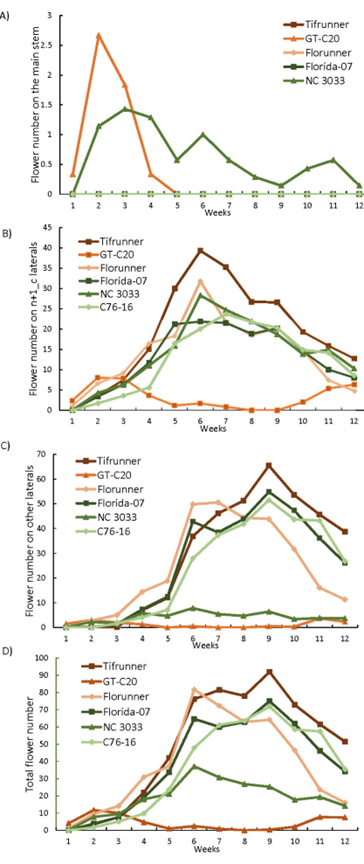
Flower counts for the 12 weeks after 11 DAT demonstrated varied flowering patterns among the tested genotypes (Figure 1). GT-C20 is a spanish-type peanut and NC 3033 has 50% spanish background in its pedigree. The flowering patterns of these two lines are expected to be distinct from the four runner type peanuts. In this experiment, GT-C20 and NC 3033 were the only two genotypes bearing flowers on the main stem (Figure 1A). GT-C20 produced flowers on the main stem on the first week of observation, peaked flower production by the second week and halted flowering on the main stem by the fifth week. NC 3033 started to flower on the main stem from the second week until the 12th week. Although both GT-C20 and NC 3033 produced flowers on the main stem, the different flower patterns between the two genotypes suggests that they may possess distinct genetic controls for the timing of flowering on the mainstem.
GT-C20 had a distinct flowering pattern on n+1_c laterals compared to the other genotypes (Figure 1B). It mainly produced flowers from weeks 2 to 5 and weeks 10 to 12 with very little flower production from weeks 6 to 9. This early and centralized flowering pattern on n+1_c contributes to the early maturity of this genotype as reported in other spanish peanut cultivars (Kaba et al., 2014). The other genotypes exhibited close to a normal distribution of flower numbers on the n+1_c laterals with peak flower production between weeks 5-8. As for other laterals (Figure 1C), both GT-C20 and NC 3033 produced very few flowers throughout the growing season. This is in contrast to the other four genotypes which exhibited a normal distribution similar to the patterns on n+1_c except that the peak production time appeared to shift a couple of weeks later.
Trend lines for total number of flowers were plotted (Figure 1D) and the largest difference was between Tifrunner and GT-C20. Statistical analysis of the total number of flowers by repeated measure indicated a significant effect of time (F=117.1, p<0.0001), genotype (F=42.7, p<0.0001), and the interaction between time and genotype (F=14.3, p<0.0001). Weekly comparison of LS (least square) mean of the flower counts among genotypes (Table 1) indicated that GT-C20 had the highest LS mean during the first two weeks and became the genotype that produced the least number of flowers from weeks 4-12. Florunner had the highest LS mean from weeks 3-7 and was replaced by Tifrunner from weeks 8-12. Statistically significant differences of total flower numbers were found in 9 out of 12 weeks between Tifrunner and GT-C20. Therefore, the population with Tifrunner and GT-C20 as parents is suitable to study total flower number and flower distribution.
Flower-to-peg conversion
Upon successful fertilization and initiation of embryo development in peanut, a meristem at the base of the ovary causes elongation of a stalk-like structure commonly known as the peg (Smith, 1950). Geotropic growth of the peg allows the formation of peanut fruits underground. Low efficiency of both the flower-to-peg and the peg-to-mature pod conversion steps negatively affect time-to-maturity and peanut productivity. It has been noted that 40 to 70% of peanut flowers formed pegs and among the elongated pegs only a fraction resulted in mature pods (Smith, 1954; Coffelt et al., 1989). Environmental factors such as temperature and photoperiod have been shown to affect flower-to-peg conversion ratio (Bagnall and King, 1991). In addition, significant genotypic variation for flower-to-peg conversion ratio was observed (Coffelt et al., 1989; Puangbut et al., 2013).
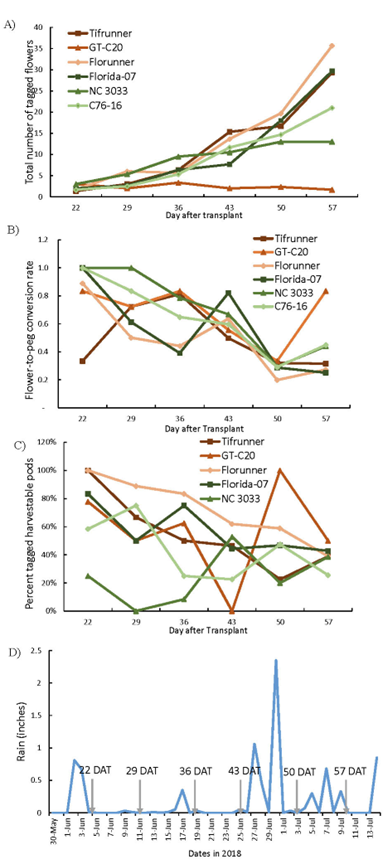
In this study, a separate set of plants from the flower count study were used to determine flower-to-peg conversion rates. Snapshots of flower-to-peg conversion were captured at six time points after 80% of the plants produced their first flower. The trends of the total number of tagged flowers at the six time points (Figure 2A) were similar to that of the total flower plots reported in the previous section (Figure 1D). There was a simultaneous drop in flower-to-peg conversion rate at 50 DAT for all tested genotypes (Figure 2B). This drop coincided with two heavy rainfall events between 43 to 50 DAT (Figure 2D). Peanut flowers open soon after sunrise and wilt in the afternoon (Smith, 1950). Wilted flowers usually remain at the base of the axil and attach to the tip of elongated pegs until soil penetration of the pegs. However, the attachment of the wilted flower is rather weak. Multiple rainfalls after flower tagging and before peg tagging may have caused detachment of tagged flowers and reduced the chance of recovering identifiable pegs.
Tagged flower number and peg-to-pod production monitored at six time points. Total number of flowers tagged at each time point (A), flower to peg conversion rate (B), percent of tagged harvestable pods over recovered tags at harvest (C). Rainfall record (D). The six time points overlapped with week 2 to week 7 of Figure 1.
Flower-to-peg conversion rates demonstrated a complex pattern (Figure 2B) with a significant effect of genotype (F=3.62, p=0.0061), time (F=18.4, p<0.0001), and time x genotype interaction (F=2.19, p=0.0063). Comparison of genotypic effect across the six-time points (Table 2) indicated that Tifrunner had a significantly lower LS mean value (0.3) than the other five genotypes at 22 DAT. No significant separation of LS means among genotypes were found from 29 to 50 DAT. At 57 DAT, GT-C20 reached a LS mean of 0.8, which was significantly higher than that of Florunner, Florida-07, and Tifrunner. At this time point, GT-C20 produced only one or two flowers, whereas the other genotypes produced an average of 10 to 35 flowers. Although the number of flowers produced by GT-C20 was small, they were fertilized and formed pegs which resulted in 50 to 100% flower to peg conversion ratios. On the other hand, the remaining genotypes made more pegs yet produced an even higher number of flowers than GT-C20 resulting in lower flower-to-peg conversion ratio than GT-C20. The overall average percent of tagged pegs decreased between 22 and 57 DAT suggesting a decrease in efficiency of peg production as the flower production increases. From the population perspective, the only significant difference of flower-to-peg conversion rate was found between Tifrunner and GT-C20 at 22 and 57 DAT. This method of tagging flowers and pegs in the field is not feasible to apply to mapping populations with large numbers of lines since this task is labor intensive and time consuming. In addition, incidences such as rainfall and animal activities that disturb the attachment of tagged flowers or cause loss of pods at harvest can confound the results.
At harvest, 392 peg tags were recovered from the total of 440 tags attached. The timeline of the percentage of R4 to R8 harvestable pods over tagged pegs (peg-to-pod conversion) was plotted (Figure 2C). The average conversion rate ranged from 0.19 to 0.65, similar to a previous report (Coffelt et al., 1989). Significant effects of genotype (F=5.24, p=0.0006) and time (F=3.52, p=0.0081) were found whereas the time x genotype interaction (F=1.64, p=0.07) was not significant. Within the six days of snap shots, the overall percentage of tagged harvestable pods of Florunner and NC 3033 were the highest (57%) and lowest (20%), respectively (Table 3). Peanut has an indeterminant growth habit and produces flowers throughout most of the growing season. These tagged pods only accounted for less than 5% of the total production of pods. As for the percentage of total R4-8 pods among total pegs at harvest, GT-C20 had the highest percentage (52%) and NC 3033 had the lowest (21%). The high percentage of harvestable pods in GT-C20 is consistent with its more determinate nature compared to other genotypes in this study. A 35% correlation was found between the percentage of tagged pods from six time points and percentage of total pods at harvest.
Although the range of pod conversion ratio is narrow, the total peg and pod production among the genotypes had a much wider span. Between the genotypes with largest difference of pod and peg production, Tifrunner produced six times more pegs than GT-C20 and C76-16 produced 4.5 times more pods than GT-C20 (Table 3). Yield of peanut is the product of total number of pegs and peg-to-pod conversion ratio. A high production of pegs in conjunction with a high peg-to-pod conversion ratio leads to high yield. Taking snap shots of peg-to-pod conversion ratio during the growing season illuminates the chronological changes of peanut productivity, however, it is prohibitive to perform this type of measurement with large populations due to labor constraints.
Morphological characteristics
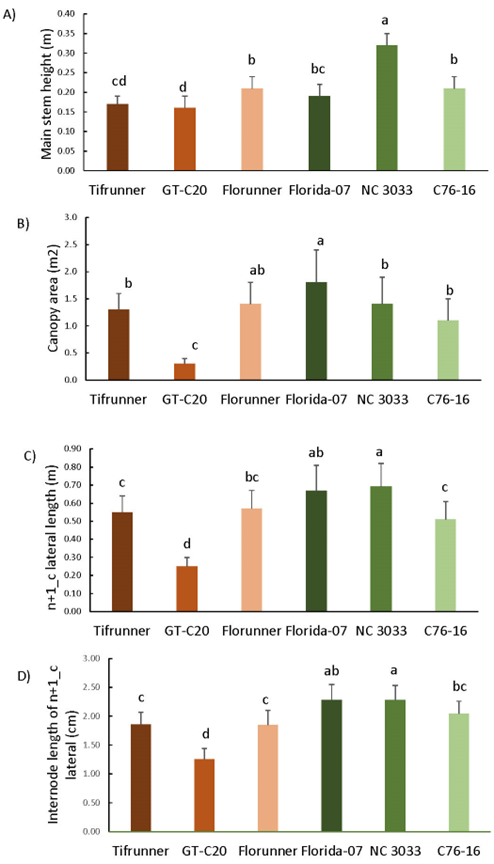
Plant morphological characteristics influence time-to-maturity by determining the architecture of peanut plants and the spatial and temporal distributions of flowers and pods. These traits include main stem (n) height, canopy area, n+1_c lateral length, n+1_c internode length, distribution of reproductive and vegetative nodes on the n+1_c laterals, and number of branches on n+1, n+2 and n+3 laterals. Significant differences were found among the tested genotypes for these morphological traits collected at harvest (Figure 3). GT-C20 was the smallest among the tested genotypes for all measurements except for percentage of reproductive nodes on n+1_c laterals. Average main stem height ranged from 0.16 m to 0.31 m with NC 3033 being the tallest (Figure 3A). Florida-07 had the largest canopy area, which was about three times the size of GT-C20 (Figure 3B). NC 3033 had the longest n+1_c laterals and internode length although it was comparable to Florida-07 (Figure 3C and 3D). As for the distribution of vegetative and reproductive nodes on the n+1_c laterals (Figure 3E and 3F), GT-C20 and NC 3033 had significantly higher percentages of reproductive nodes than Tifrunner, Florunner, and C76-16. For the alternating frequency of vegetative and reproductive nodes, GT-C20 was the lowest followed by NC 3033, whereas Tifrunner had the numerically highest alternating frequency. These data conform with the distinction between the spanish versus runner types in which spanish peanut had consecutive reproductive nodes and runner had alternating pairs of vegetative and reproductive nodes on the n+1_c laterals (Wynne, 1975).
Morphological traits collected at harvest including main stem height (A), canopy area (B) n+1_c lateral length (C), internode length of n+1_c laterals (D), percentage of reproductive nodes on n+1_c laterals (E), alternating frequency between V- and R- nodes on n+1_c laterals (F), number of branches of n+1, n+2 and n+3 laterals (G). Bars above data columns represent standard error. Different letters on top of the bars indicate the statistically significant differences in grouping at P<0.05.
Number of n+1 lateral branches was the lowest compared to numbers of n+2 and n+3 lateral branches for most genotypes (Figure 3G). Florunner and Tifrunner had the highest number of n+1 lateral branches with an average of 15. GT-C20, the genotype with the lowest branch number, had an average of seven n+1 lateral branches. As for number of n+2 and n+3 laterals among Florida-07, Florunner, Tifrunner, C76-16, and NC 3033, the only significant difference was found in n+2 branch number of NC 3033. However, these five genotypes had significantly higher numbers of n+2 and n+3 lateral branches than GT-C20. The average numbers of n+2 and n+3 laterals for Tifrunner were 56 and 77, respectively, whereas GT-C20 only had 16 and five branches.
Yield related traits
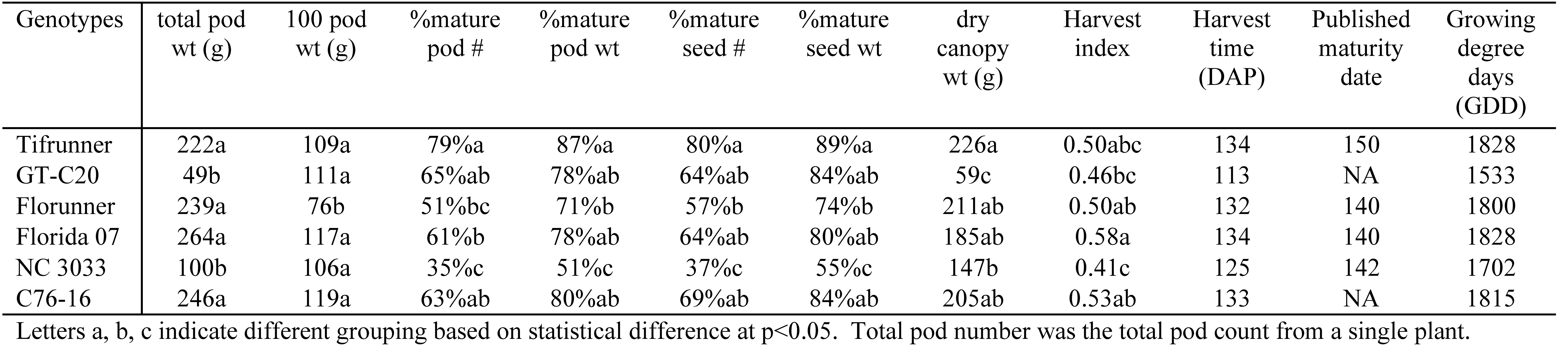
Upon harvest, several yield related traits were measured including R2-R3 peg number, R4-R8 pod number, total peg number, 100 double pod weight, dry canopy weight, total pod weight, percent of mature pods, percent of mature seeds and harvest index. The first three measurements were reported in Table 3 in the previous section to make a comparison with the results from tagged pegs. The remaining measurements (Table 4) indicated that total pod weight (pod yield) for Florunner, Tifrunner, C76-16, and Florida-07 were significantly higher than both NC 3033 and GT-C20. Total pod yield per plant of Florida-07, the highest yielding line, was 264 grams and GT-C20 only yielded 49 grams. Florunner had the lowest 100 pod weight and was significantly lower than the rest of the genotypes. Florunner was a predominant cultivar grown in the southeastern U.S. until the appearance of TSWV (Culbreath and Srinivasan, 2011). Its high level of susceptibility to TSWV made it unsuitable for cultivation in this region. However, its small and round seed size, shape and flavor is highly desirable for the peanut industry. Peanut size is larger in most of the recently released cultivars. Although increased seed size can contribute to yield, breeding for preferred size peanut is important to meet the demand of the peanut industry. Percent of mature pod/seed number and weight correlates with the estimation for optimal harvest time. We harvested each genotype at multiple time points based on pod maturity levels determined by the hull scrape method which were earlier than published maturity dates. The discrepancy could be caused by the limited number of plants and pods available for sampling. Tifrunner reached 80% of mature pods suggesting that the timing for harvest for this line was close to optimum. NC 3033 had the lowest percentage of mature pods (35%) indicating that the harvest time of this line was too early. Dried canopy weight of Tifrunner was the highest and GT-C20 was the lowest. As for harvest index, the ratio of pod yield versus pods plus canopy weight, Florida-07 was the highest and NC 3033 was the lowest. Growing degree days (GDD) measures the cumulative average daily temperature above the base temperature arresting peanut growth during the growing season. It has been found that GDD is associated with physiological maturity of peanut plants and a model of using GDD to predict peanut maturity was established (Rowland et al., 2006). Recently released virginia cultivars tend to be early maturing and require lower GDD to reach physiological maturity than older cultivars (Oakes et al., 2020). GDD of the tested genotypes in this study were comparable to the reported GDD of peanut cultivars Bailey, Sullivan and Wynne (Oakes et al., 2020).
Previously we identified nine QTL for pod and seed size on chromosomes A05, A06, A09, B10, B04, A03, B05, and B08 using Florida-07 x GP NC WS 16 RIL population (Chu et al., 2019). The major QTL on chromosome A05 explains 66% of phenotypic variation. In addition, Tifrunner x NC 3033 population was mapped for seed size index, kernel percentage, seed weight, pod weight, single and double kernel, pod area and density (Chavarro et al., 2020). Consistent QTL for kernel percentage were identified on chromosomes A07/B07. Major QTL for pod and seed weight were found on chromosome B06. Additional mapping efforts for this trait may result in new QTL influencing these traits.
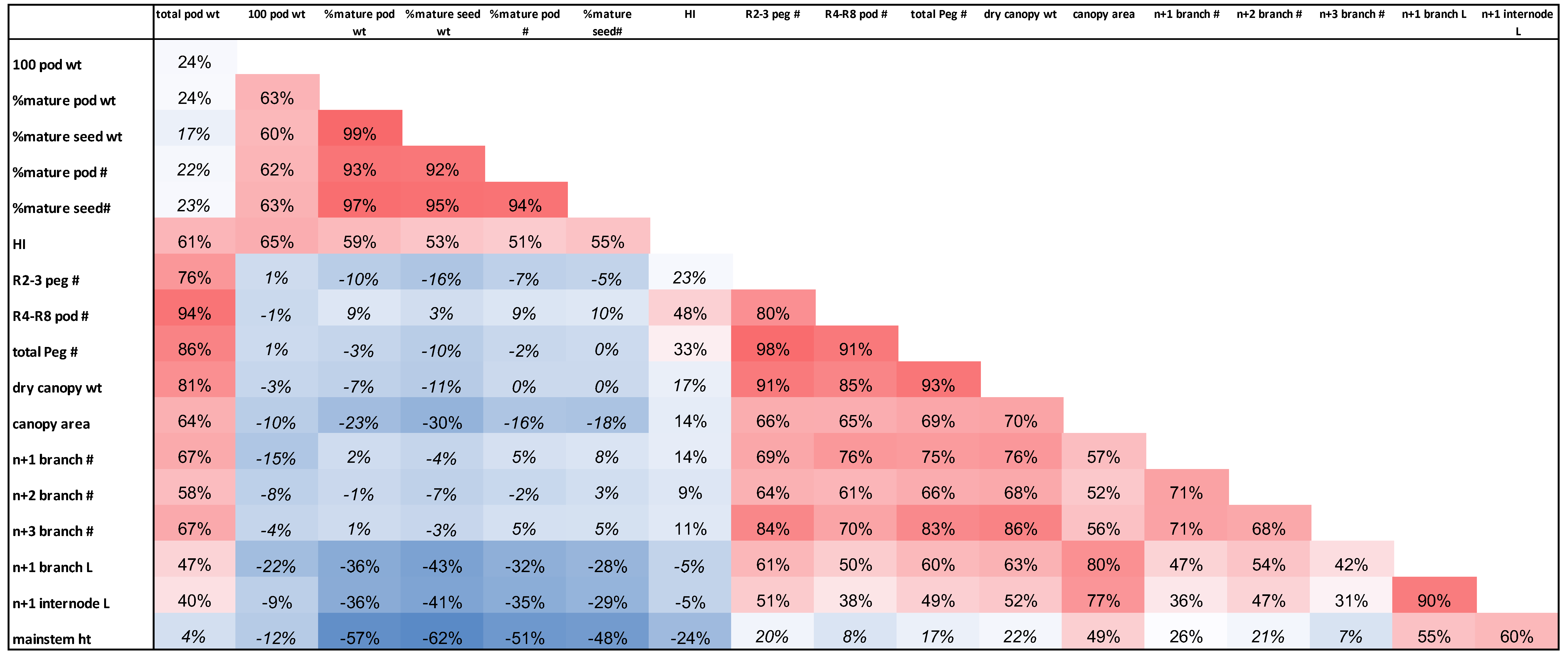
Pearson correlation analysis was performed with all of the morphological and yield related traits collected from these genotypes (Table 5). Most of the traits demonstrated significant correlations among each other. In this study, R4-R8 pod number, total peg number, dry canopy weight, canopy area, harvest index, branch numbers, branch length, 100 pod weight and percent of mature pod weight were significantly positively correlated with pod yield. Particularly, R4-R8 pod number, total peg number and dry canopy weight had a correlation greater than 80% to pod yield suggesting the high level of impact of these traits on pod yield. Although percent of mature pod weight and number, percent of mature seed weight and number had a low percentage of correlation to pod yield, high percentage of mature pod and seed at harvest directly improves seed quality, reduces moisture content and subsequently post-harvest aflatoxin contamination. Most of peanuts are processed by the food industry for human consumption. The quality of peanut in this respect is defined by roast color, flavor and storability (Sanders 1989; Sanders et al., 1989). Mature peanut was found to have the desirable roasted peanutty and sweet aromatic flavor as opposed to the bitter and painty taste of immature peanuts (Sanders et al., 1989). On the other hand, a portion of the peanuts are allocated as seed stock for farmers in which peanut quality is determined by the rate of seed germination and seedling vigor (Lamb et al., 1997). Mature peanuts were shown to have higher germination rates and seedling vigor than immature peanuts (Dey et al., 1999). Therefore, both improving yield and uniformity of peanut maturity need to be simultaneously considered for cultivar improvement. In addition, total pod weight was positively correlated with lateral branch number and length, canopy weight and size suggesting these measurements of peanut plant architecture may contribute to yield.
Current advancements in peanut genetics and genomics allow for construction of high-density genetic maps by exploring single nucleotide polymorphisms among genotypes (Clevenger et al., 2015; Bertioli et al., 2019). The bottleneck of genetic mapping becomes the acquisition of phenotypic data. The indeterminate growth habit of peanut poses a great challenge in determining time-to-maturity. To circumvent this obstacle, we investigated component traits contributing to time-to-maturity in peanut. GT-C20 and NC 3033, the two lines with spanish background, demonstrated clear phenotypic differences from the runner type peanuts. Phenotypic differences among population parents provide guidelines to choose the appropriate population for further genetic mapping analysis.
Acknowledgements
The authors would like to express their appreciation for the statistical support from Ms. Xuelin Luo and technical support from Stephanie Botton, Shannon Atkinson, and Jason Golden. This research was supported by the US-Israel Binational Agricultural Research and Development Fund (BARD US-5020-17) to POA and RH and the Peanut Research Foundation.
Literature Cited
Ali N., and Wynne J. C.. 1994. Heritability estimates and correlation studies of early maturity and other agronomic traits in two crosses of peanut (Arachis hypogaea L.). Pak. J. Bot. 26: 75-82.
Bagnall D. J., and King R. W.. 1991. Response of peanut (Arachis hypogea) to temperature, photoperiod and irradiance effect on peg and pod development. Field Crops Res. 26: 279-293.
Bertioli D. J., Jenkins J., Clevenger J., Dudchenko O., D. Y. Gao et al. 2019. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 51: 877-884.
Beute M. K., Wynne J. C., and Emery D. A.. 1976. Registration of NC 3033 peanut germplasm. Crop Sci. 16.
Boote K. J. 1982. Growth stages of peanut (Arachis hypogaea L.). Peanut Sci. 9: 35-40.
Chavarro C., Chu Y., Holbrook C. C., Isleib T., Bertioli D., Hovav R., Butts C., Lamb M., Sorensen R., Jackson A. S., and Ozias-Akins P.. 2020. Pod and seed trait QTL identification to assist breeding for peanut market preferences. G3 10:2297-2315.
Chirinos F. V. 2011. Breeding for early maturity in peanuts (Arachis hypogaea L.) using traditional methods and marker assisted selection (MAS), pp. 120. North Carolina State University, Raleigh, NC.
Choudhari S. D., Udaykumar M., and Sastry K. S. K.. 1985. Physiology of bunch groundnuts (Arachis hypogaea L.). J. Agri. Sci. 104: 309-315.
Chu Y., Chee P., Isleib T. G., Holbrook C. C., and Ozias-Akins P.. 2019. Major seed size QTL on chromosome A05 of peanut (Arachis hypogaea) is conserved in the U.S. mini core germplasm collection. Mol. Breed 40:6
Chu Y., Holbrook C. C., Isleib T. G., Burow M., A. K. Culbreath et al. 2018. Phenotyping and genotyping parents of sixteen recombinant inbred peanut populations. Peanut Sci. 45: 1-11.
Clevenger J., Chavarro C., Pearl S. A., Ozias-Akins P., and Jackson S. A.. 2015. Single nucleotide polymorphism identification in polyploids: A review, example, and recommendations. Mol. Plant 8: 831-846.
Coffelt T. A., Seaton M. L., and VanScoyoc S. W.. 1989. Reproductive efficiency of 14 Virginia-type peanut cultivars. Crop Sci. 29: 1217-1220.
Culbreath A. K., and Srinivasan R.. 2011. Epidemiology of spotted wilt disease of peanut caused by Tomato spotted wilt virus in the southeastern U.S. Virus Res 159:101-109.
Dey G., and Mukherjee R. K., Bal S.. 1999. Influence of harvest and post-harvest conditions on the physiology and germination of peanut kernels. Peanut Sci. 26:64-68.
Giayetto O., Morla F. D., Fernandez E. M., Cerioni G. A., Kearney M. et al., 2013. Temporal analysis of branches pod production in peanut (Arachis hypogaea) genotypes with different growth habits and branching patterns. Peanut Sci. 40: 8-14.
Gilman D. F., and Smith O. D.. 1977. Internal pericarp color as a subjective maturity index for peanut breeding. Peanut Sci 4:67-70.
Gorbet D. W., and Tillman B. L.. 2008. Registration of 'Florida-07' peanut. J. Plant Reg. 3: 14-18.
Holbrook C. C., Burow M. D., Chen C. Y., Pandey M. K., Liu L. et al. 2016. Recent advances in peanut breeding and genetics, pp. 111-145 in Peanuts: Genetics, Processing, and Utilization., edited by H. T. Stalker and R. F. Wilson. AOCS Press, Elsevier Inc.
Holbrook C. C., Cantonwine E., Sullivan D. G., Guo B., Wilson D. M. et al. 2007. Development of peanut germplasm with improved drought tolerance, pp. 21 in Proc. Amer. Peanut Res. and Educ. Soc.
Holbrook C. C., and Culbreath A. K.. 2007. Registration of ‘Tifrunner’ peanut. J. Plant Reg. 1: 124.
Holbrook C. C., Isleib T. G., Ozias-Akins P., Chu Y., S. Knapp et al. 2013. Development and phenotyping of recombinant inbred line (RIL) populations for peanut (Arachis hypogaea). Peanut Sci. 40: 89-94.
Holbrook C. C., and Stalker H. T.. 2002. Peanut breeding and genetic resources, pp. 297-356 in Plant Breeding Reviews, edited by J. Janick. John Wiley & Sons, Inc.
Hong Y., Chen X., Liang X., Liu H., G. Zhou et al. 2010. A SSR-based composite genetic linkage map for the cultivated peanut (Arachis hypogaea L.) genome. BMC Plant Biol. 10: 17.
Huyghe C. 1998. Genetics and genetic modifications of plant architecture in grain legumes: a review. Agron. EDP Sci. 18: 383-411.
Kaba J. S., Kumaga F. K. and Ofori K.. 2014. Effect of flower production and time of flowering on pod yield of peanut (Arachis hypogaea L) genotypes. IOSR-JAVS 4: 44-49.
Krapovickas A., and Gregory W.C.. 2007. Taxonomy of the genus Arachis (Leguminosae). Translated by D. E. Williams & C. E. Simpson. Bonplandia 16 (supl.): 1-205.
Kunta S., Agmon S., Hedvat I., Levy Y., Chu Y., Ozias-Akins P., and Hovav R.. 2021. Identification of stable QTL for time to mature in Virginia-type peanut (Arachis hypogaea L.). BMC Plant Biol. DOI: https://doi.org/10.21203/rs.3.rs-200335/v1
Kvien C. K., and Ozias-Akins P.. 1991. Lack of monocarpic senescence in Florunner peanut. Peanut Sci. 18: 86-90.
Lamb M. C., Davidson J. I., Childre J. W., and Martin N. R.. 1997. Comparison of peanut yield, quality, and net returns between nonirrigated and irrigated production. Peanut Sci 24:97-101.
Lamb M. C., Sorensen R. B., Nuti R. C., Rowland D. L., W. H. Faircloth et al. 2010. Impact of sprinkler irrigation amount on peanut quality parameters. Peanut Sci. 37: 100-105.
Liang X. Q., Luo M., Holbrook C. C., and Guo B. Z.. 2006. Storage protein profiles in Spanish and runner market type peanuts and potential markers. BMC Plant Biol. 6: 24.
Nigam S. N., and Aruna R.. 2008. Improving breeding efficiency for early maturity in peanut. Plant Breeding Rev. 30: 295-322.
Norden A. J., lipscomb R. W., and Carver W. A.. 1969. Registration of Florunner peanut. Crop Sci 9: 850.
Oakes J. C., Balota M., Jordan D. L., Hare A. T., and Sadeghpour A.. 2020. Peanut response to seeding density and digging date in the Virginia-Carolina region. Peanut Sci. 47:180-188
Pattee H. E., Johns E. B., Singleton J. A. and Sanders T. H.. 1974. Composition changes of peanut fruit parts during maturation. Peanut Sci. 1: 57-62.
Puangbut D., Jogloy S., Vorasoot N., Kesmala T., C. C. Holbrook et al. 2013. Response of reproductive parts of peanut genotypic variation and their contributions to yield after pre-flowering drought AJCS 7: 1627-1633.
Qin H., Feng S., Chen C., Guo Y., Knapp S., Culbreath A., He G., Wang M. L., Zhang X., Holbrook C. C., Ozias-Akins P., and Guo B.. 2012. An integrated genetic linkage map of cultivated peanut (Arachis hypogaea L.) constructed from two RIL populations. Theor. Appl. Genet. 124:653-664.
Rife T. W., and Poland J. A.. 2014. Field Book: An open-source application for field data collection on android. Crop Sci. 54: 1624-1627.
Rowland D. L., Sorensen R. B., Butts C. L., and Faircloth W. H.. 2006. Determination of maturity and degree day indices and their success in predicting peanut maturity. Peanut Sci. 33:125-136.
Rucker K.S., Kvien C.K., Calhoun K., Henning R.J., Koehler P.E., Ghate S.R., et al. 1994. Sorting peanuts by pod density to improve quality and kernel maturity distribution and to reduce aflatoxin. Peanut Sci. 21: 147-152.
Sanders T. H. 1989. Maturity distribution in commercially sized Florunner peanuts. Peanut Sci. 16:91-95.
Sanders T. H., Vercellotti J. R., Crippen K. L., and Civille G. V.. 1989. Effect of maturity on roast color and descriptive flavor of peanuts. J. Food Sci. 54:475-477.
Shashidhar V. R., Chari M., Prasad T. G., and Udaya K. M.. 1986. A physiological analysis of the branching pattern in sequential types of groundnut in relation to the fruiting nodes and the total mature pods produced. Ann. Bot. 58: 801-807.
Smith B. W. 1950. Arachis hypogaea aerial flower and subterranean fruit. Am. J. Botany 37: 802-815.
Smith B. W. 1954. Arachis hypogaea. Reproductive efficiency. Am. J. Botany 41: 607-616.
Sorensen R. B., Butts C. L., and Lamb M. C.. 2020. X-ray technology to determine peanut maturity. Peanut Sci. 47: 38-45.
Stalker H. T. 2017. Utilizing wild species for peanut improvement. Crop Sci. 57: 1102-1120.
Stalker H. T., and Simpson C. E. 1995. Germplasm resources in Arachis. In: Pattee HE, Stalker HT (eds) Advances in peanut science. American Peanut Research and Education Society, Inc, Stillwater, OK, pp 14-53.
Wang H., Pandey M. K., Qiao L. X., Qin H. D., Culbreath A. K. et al. 2013. Genetic mapping and quantitative trait loci analysis for disease resistance using F2 and F5 generation-based genetic maps derived from 'Tifrunner' x 'GT-C20' in peanut. Plant Genome 6. Available at: DOI: 10.3835/plantgenome2013.05.0018
Williams E. J., and Drexler J. S.. 1981. A non-destructive method for determining peanut pod maturity. Peanut Sci. 8: 134-141.
Wynne J. C. 1975. Inheritance of branching pattern in Arachis hypogaea L. Peanut Sci. 1: 1-5.
Notes
-
Author Affiliations
- Horticulture Department, University of Georgia, Tifton, GA, USA. [^]
- USDA- Agricultural Research Service, Crop Genetics and Breeding Research Unit, Tifton, GA, USA. [^]
- Department of Vegetables & Field Crops Research, Institute of Plant Sciences, Agriculture Research Organization-the Volcani Center, HaMakkabbim Road, P. O. Box 15159, 7505101 Rishon LeZiyyon, Israel. [^]
- Institute of Plant Breeding, Genetics and Genomics, University of Georgia, Tifton, GA, USA. [^] Corresponding author: Peggy Ozias-Akins pozias@uga.edu.