Introduction
Peanut rust, a foliar disease caused by Puccinia arachidis Speg., is a widespread problem in countries with warm, tropical climates. The disease reduces peanut yield and quality and indirectly increases management costs (Subrahmanyam et al., 1997). Rust often co-occurs with early leaf spot (caused by Passalora arachidicola (Hori) U. Braun [syn. Cercospora arachidicola (S. Hori)]), and late leaf spot (caused by Nothopassalora personata (Berk. & M.A. Curtis) U. Braun, C. Nakash, Videira & Crous [syn. Cercosporidium personatum (Berk. & M. A. Curtis) Deighton]), which have been reported to cause up to 80% yield losses in India in the absence of fungicide control (Subrahmanyam et al., 1984). Rust is primarily a pathogen that afflicts tropical countries and has only sporadic outbreaks in the U.S. However, global climate change may increase the impact of this disease through increasing the frequency of tropical storms that carry rust inoculum into the U.S. and by expanding the range in which rust can overwinter (Power, 2014). Yield loss estimates due to rust alone are unavailable for the U.S. due to it being a localized issue in warm regions and its co-occurrence with leaf spots; however, losses due to damage caused by rust and early and late leaf spot and increased fungicide costs were approximately $32 million in Georgia in 2011 (Williams-Woodward, 2013). Crop rotations, eradicating volunteer peanut plants to reduce inoculum source, and allowing one-month fallow periods are cultural practices applied by farmers who do not have access to or cannot afford fungicides in countries such as India, Haiti, and Guyana (Subrahmanyam, 1997). Even for farmers that can use fungicides, there is still a likelihood that rust populations will develop resistance when exposed to frequent fungicide applications (Smith and Littrell, 1980). Therefore, rust resistant, high-yielding peanut cultivars are an important part of an integrated pest management strategy to control rust in tropical countries as well as the U.S.
A major limitation to breeding rust-resistant cultivars is that only moderate levels of resistance have been identified in A. hypogaea germplasm (Power et al., 2019). Fortunately, high resistance to rust has been identified in numerous wild Arachis species, including the readily usable A- and B-genome Arachis species, and resistance can be introgressed into cultivated peanut (Subrahmanyam et al., 1982, 1983; Pande and Rao, 2001; Fávero et al., 2009). Cultivated peanut is an allotetraploid (AABB; 2n=4x=40) species and the majority of wild Arachis species are diploid (2n=2x=20); the most efficient way to introgress genes from wild Arachis species into cultivated peanut is to cross A- and B-genome species to produce allotetraploid interspecific hybrids that are cross-compatible to peanut. Then, the wild Arachis derived allotetraploids are backcrossed to peanut cultivars. Markers linked to rust resistance allow quick introgression as well as pyramiding of multiple QTLs. Rust resistant QTLs have been identified in the A-genome species A. cardenasii GKP 10017 and the B-genome species A. magna K 30097. One major QTL from each species is being used in peanut breeding programs to introgress rust resistance into peanut cultivars (Khedikar et al., 2010; Sujay et al., 2012; Leal-Bertioli et al., 2015). For example, Gowda et al. (2002) released a rust resistant cultivated genotype ‘GPBD 4] (ssp. fastigiata var. vulgaris) with bunch growth habit, in which resistance was derived from A. cardenasii. GPBD 4 had a mean rust score of three on a scale of one to nine, in which one was equivalent to no disease and nine was equivalent to 80 to 100% disease (Gowda et al., 2002). While GPBD 4 still develops rust, it does so at far lower levels than most cultivated germplasm, and one rust resistance QTL derived from A. cardenasii but identified in a population derived from GPBD 4, has been shown to improve yields by 56 to 96% in rust infected environments (Gowda et al., 2002; Varshney et al., 2014). While progress towards resistant rust cultivars is being made, more major rust resistance QTLs need to be identified for further pyramiding in peanut cultivars to strengthen resistance and more importantly, to increase resistance durability to rust population pressures. This study identified rust resistance in newly synthesized allotetraploids that are cross compatible with cultivated peanut. The long-term goal of this study is to create rust resistant peanut cultivars that can protect yields and decrease the need for fungicides in the U.S. and to increase yields in tropical, developing countries for farmers who cannot afford or access costly fungicides.
Materials and Methods
Plant Materials
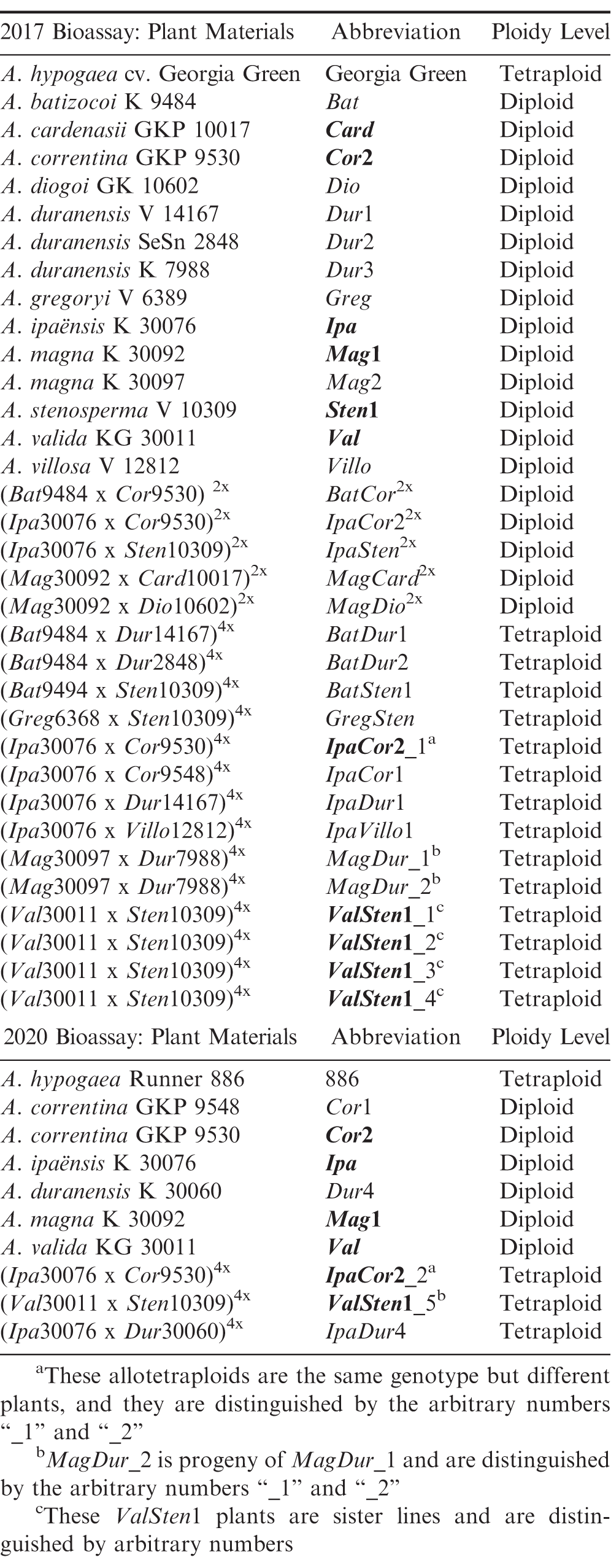
Diploid, wild Arachis species, A. correntina (Burkart) Krapov. and W.C. Gregory [PI 262808, GKP 9530 (Cor9530)], A. duranensis Krapov. and W.C. Gregory [PI 468197, GKBSPSc 30060 (Dur)], and A. ipaënsis Krapov. and W.C. Gregory [PI 468322, GKBSPSc 30076 (Ipa)] were used to generate the diploid hybrids, IpaCor2 and IpaDur4 in 2016 at North Carolina State University (NCSU). Wild Arachis species, A. batizocoi Krapov. and W.C. Gregory [PI 298639, K 9484 (Bat)], A. cardenasii Krapov. and W.C. Gregory [PI 261874, GKP 10017 (Card)], A. correntina (Cor9530) and [PI 262881, GKP 9548 (Cor9548)], A. diogoi [PI 331200, GK 10602 (Dio)], A. duranensis Krapov. and W.C. Gregory [V 14167 (Dur1), SeSn 2848 (Dur2), and K 7988 (Dur3)], A. gregoryi A. Gripp, C.E. Simpson, and J.F.M. Valls [PI 476116, VSGr 6389 (Greg)], A. magna Krapov., W.C. Gregory, and C.E. Simpson [PI 468337, K 30092 (Mag1) and PI 468340, K 30097 (Mag2)], A. stenosperma Krapov. and W.C. Gregory [V 10309 (Sten1) and PI 338280, HLK 410 (Sten2)], A. valida Krapov. and W.C. Gregory [PI 468154, KG 30011 (Val)] and A. villosa Benth. [V 12812 (Villo)] were used to create diploid hybrids BatCor, BatDur1, BatDur2, BatSten1, GregSten, IpaCor1, IpaDur1, IpaSten, IpaVillo1, MagCard, MagDio, MagDur, and ValSten1 at the University of Georgia (UGA) Athens Campus. All allotetraploids were derived from the diploid hybrids by colchicine treatment of F1 hybrid cuttings at the UGA Athens Campus, except for IpaDur4 and IpaCor2, which were generated at the UGA Tifton Campus.
The resistance evaluation was performed in two separate experiments, in which four wild Arachis species and two interspecific hybrid combinations, IpaCor2 and ValSten1, were tested in both experiments (Table 1). Allotetraploids that had more than one plant tested were each designated an arbitrary number to make distinguishing them easier (Table 1). The first experiment performed in 2017 included A. hypogaea ‘Georgia Green’ (Branch, 1996) as a susceptible control, while the 2020 experiment included A. hypogaea ‘Runner 886] (abbrev.: 886) as a susceptible control due to seed availability.
Rust Resistance Evaluation
For both experiments, seeds were coated in Captan + pentachloronitrobenzene + carboxin (Vitavax PC, Vitavax, Crompton, Middlebury, CT) and treated overnight in 0.5% ethephon [(2-chloroethyl) phosphonic acid] (Florel Growth Regulator, Lawn and Garden Products Inc., Fresno, CA) to break dormancy. Seeds were then planted in #123 7.62 cm round x 11.43 cm deep Jiffy Pots (Harris Seeds, Rochester, NY) and transplanted approximately one month later into 121.92 cm round x 27.94 cm deep pots filled with Promix growth medium (Premier Tech Horticulture, Quakertown, PA). Normal plant management was applied in the greenhouse except that fungicide treatments were withheld. One wk before each experiment, leaves infected with rust were collected from untreated border rows in Tifton, GA and rust spores were collected in sterile vials using a vacuum pump. Care was taken to avoid collecting late leaf spot spores. Spores were kept at 4 C until the day of inoculation.
For the 2017 experiment, seven newly and fully expanded leaves were collected from primary laterals from one plant per genotype (Table 1). Each leaf was washed, and then its petiole was cut diagonally underwater. The petiole was then wrapped in sterilized, water-soaked cotton and the leaf was placed in a 100 mm x 15 mm petri dish (ThermoFisher Scientific) with the abaxial side upwards. Each sterilized petri dish contained a 76 mm x 25 mm x 1 mm microscope slide (ThermoFisher Scientific) on top of a sheet of 9 cm diameter Whatman No. 1 filter paper (ThermoFisher Scientific) supported by cotton wool that was saturated with approximately 4 ml of deionized water. The cotton-wrapped petiole was in contact with the wet filter paper, while the leaflets were positioned on top of the microscope slide to avoid their contact with the wet filter paper following the method of Guimaraes et al. (2017). Mounted leaves were inoculated with a spore suspension of 0.005% Tween 20 at 1.5 × 105 urediniospores/mL of P. arachidis using a soft paint brush. Inoculated leaves were kept in the dark for 48 hr at approximately 26 C, after which they were incubated with a photoperiod of 16-hr light and 8-hr dark. The leaves were checked daily for newly emerged rust pustules to document incubation period. Susceptibility was evaluated 25 d after inoculation using the following parameters: total number of lesions/ LA(cm2) (TLA), number of sporulated lesions/leaf area (cm2) (SLA), and susceptibility index /LA (cm2) (IA) as described by (Leal-Bertioli et al., 2015). IA was calculated with the scale of Savary et al. (1989), with the following modifications made by Leal-Bertioli et al. (2015): index was the number of lesions times a number that reflected lesion size/reaction. I = Σ(s * n)/LA, where s = lesion size (1 = necrotic aborted lesion, 2−6 = ruptured, sporulating pustules, varying between 0.5 and 3 mm in diameter), n = number of lesions of a particular size, LA = leaf area (cm2). Spore count and classification was performed with a stereoscope microscope, and leaf area was measured by scanning the leaves and then using Assess 2.0 (APS Press) for image analysis. The 2020 experiment was performed the same as the 2017 experiment, except 30 replications per genotype were tested, leaves were excised from five plants per genotype, mounted leaves were inoculated with a spore suspension of 0.025% Tween 20 (ThermoFisher Scientific, Waltham, MA) at 1.7 × 106 urediniospores/mL, and the experiment was ended 28 d after inoculation.
Statistical Analysis
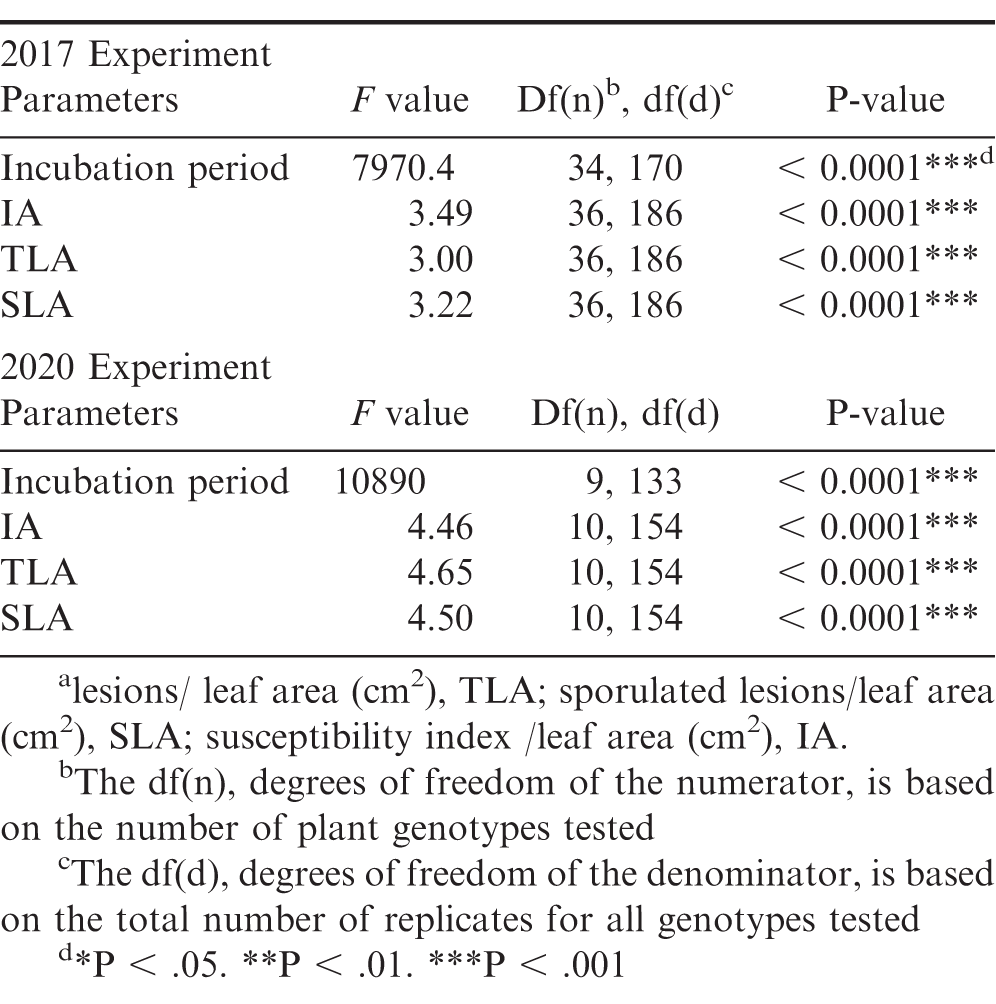
One-way analysis of variance (ANOVA) was performed using R (R Core Team, 2021) in RStudio (RStudio, Inc.) using the package agricolae (de Mendiburu, 2021) to determine the genotype effect on rust resistance according to the following parameters: incubation period, IA, TLA, and SLA. Means of each parameter among the genotypes were separated based on the Tukey’s Test (α = 0.05) results with RStudio. Greg and IpaDur1 were excluded from incubation period analysis in the 2017 experiment because each had only one replication to develop rust pustules. Genotypes that presented no rust symptoms, and therefore had no incubation period, were artificially tabulated as 100 d after inoculation for statistical analysis.
Results
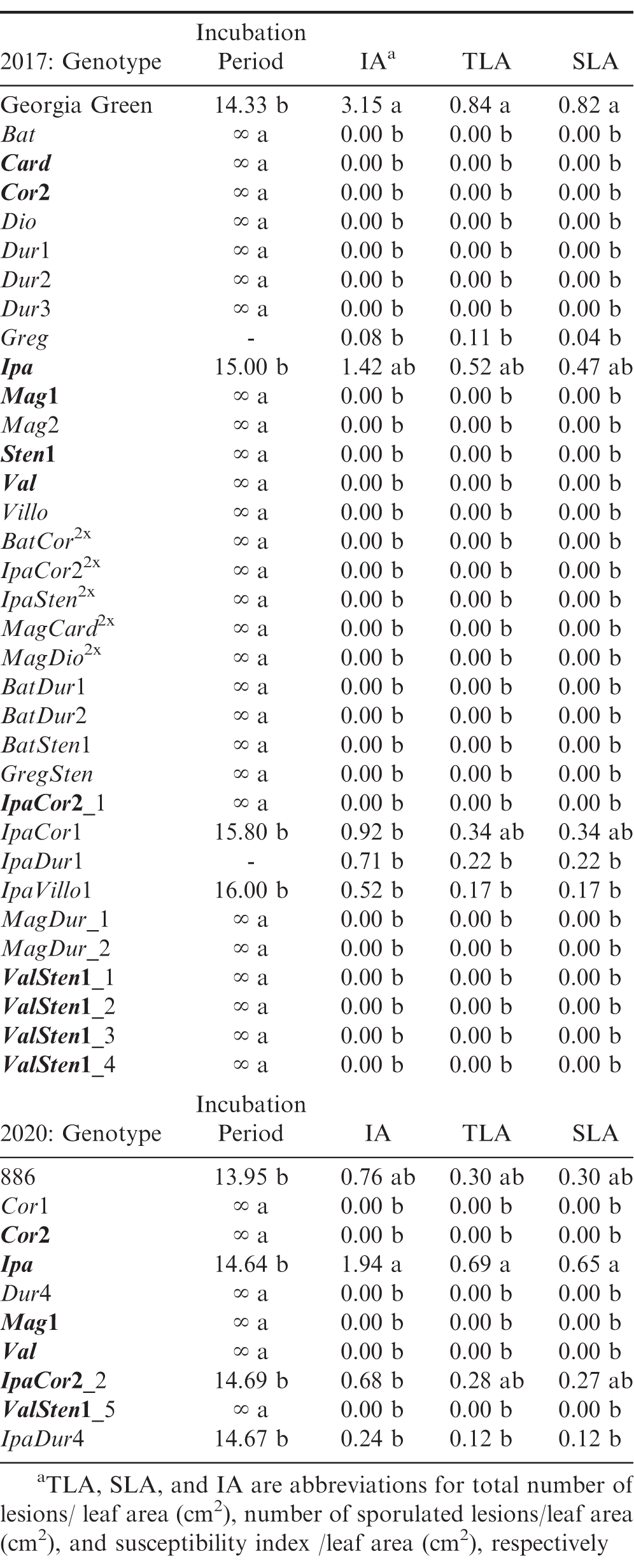
Significant genotypic effect on all rust resistance parameters was found for the 2017 and 2020 experiments (Table 2). Between both rust resistance experiments, 30 out of the 39 unique materials did not show rust symptoms and had no lesions at the end of each experiment (Table 3). In the 2017 experiment, the susceptible control, two wild Arachis species, Greg and Ipa, and three allotetraploids, IpaCor1, IpaDur1, and IpaVillo1, developed rust pustules. Greg and IpaDur1 only had one replication that developed rust pustules, so they were excluded from ANOVA analysis evaluating genotype effect on incubation time; however, these replications developed sporulating pustules, meaning they did not show a hypersensitive response. The susceptible cultivated control, Georgia Green, had the shortest incubation period of 14.33 d, while Ipa, IpaCor1, and IpaVillo1 had incubation periods within 15 to 16 d (Table 3). In the 2020 experiment, the susceptible control, one wild Arachis species, Ipa, and two allotetraploids, IpaCor2_2 and IpaDur4, developed rust pustules. The susceptible cultivated control 886 also had the shortest incubation period of 13.95 d, while Ipa, IpaCor2_2, and IpaDur4 had incubation periods between 14 and 15 d (Table 3).
Genetic materials tested in each rust bioassay, their mean for the rust resistance parameters, IA, TLA, and SLAa, and their Tukey’s HSD level. Tukey’s HSD significance levels were calculated within each experiment, so these significance groupings cannot be compared between the two experiments. Genotypes within an experiment with the same Tukey’s HSD letter are not significantly different (α = 0.05). Bolded genotypes were tested in both bioassays.
In 2017 rust experiment, the susceptible cultivated control Georgia Green had the highest IA score, but Ipa, with an IA score of 1.42, was not significantly different from Georgia Green (Table 3). IpaCor1, IpaDur1, and IpaVillo1 had IA scores of 0.92 or less, which were not significantly different from the highly resistant genotypes that did not develop rust pustules (Table 3). In the 2020 experiment, Ipa had the highest IA score of 1.94, which was significantly greater than all tested genotypes except the susceptible control, 886, which had an IA score of 0.76.
As expected, the susceptible control had the highest IA, TLA, SLA in the 2017 experiment. However, Ipa and IpaCor1 had similar TLA and SLA scores compared to the susceptible control. In the 2020 experiment, Ipa demonstrated greater susceptibility to rust with a TLA and SLA score twice as much as the susceptible control, although this difference was not significant. IpaCor2_2 showed a TLA and SLA comparable to the susceptible control. While IpaCor2_2 demonstrated susceptibility in the 2020 experiment, IpaCor22x had no rust incidence in the 2017 experiment and IpaCor2_1 had no rust incidence in the 2017 experiment.
Discussion
All the wild peanut-derived allotetraploid genotypes that were not produced with A. ipaënsis as a parent showed promise as sources for rust resistance for peanut breeding programs, since they were highly resistant to rust, with no evidence of disease development in any assay. Therefore, these nine interspecific hybrids, BatCor, BatDur1, BatDur2, BatSten1, GregSten, MagCard, MagDio, MagDur, and ValSten1 made from 13 unique Arachis accessions are recommended to peanut breeding programs for rust resistance introgression. BatSten1 and ValSten1 were deposited in the USDA-ARS National Plant Germplasm System (Fort Collins, CO) and in the USDA Plant Genetic Resources and Conservation Unit (Griffin, GA), and seed is available for research purposes (Bertioli et al., 2021; Chu et al., 2021). The other allotetraploids will be deposited for public use after sufficient disease and insect resistance characterization has been completed.
IpaCor2 performed variably in the two experiments. IpaCor22x and IpaCor2_1 had no rust incidence in the 2017 experiment, while IpaCor2_2 demonstrated susceptibility similar to the cultivated peanut control in the 2020 experiment. This variability in resistance of IpaCor2 may be due to a high level of heterogeneity in A. correntina 9530 complementing results Levinson et al. (2020) found, that different A. correntina 9530 plants and allotetraploid lines derived from this wild Arachis species accession exhibited various levels of resistance to fall armyworm in detached leaf assays. Three A. correntina 9530 plants were genotyped with the Affymetrix Axiom_Arachis2 SNP array (Clevenger et al., 2018; Korani et al., 2019) and 1,259 out of 5,342 total markers (23.5%) were found to be polymorphic (Levinson et al., 2020). This was a high level of heterogeneity when compared to A. ipaënsis and A. duranensis, which had 23 (0.4%) and 27 (0.5%) polymorphic markers, respectively (Levinson et al., 2020). The IpaCor2 plants tested in this study were made from crosses between A. ipaënsis and different A. correntina 9530 plants, so genetic difference between these plants may be due to accession heterogeneity or genetic segregation.
Of the wild Arachis species tested, only one species, Ipa, the B-genome progenitor of peanut, was susceptible to rust. In the 2020 experiment, Ipa showed higher susceptibility than the susceptible control, 886, with greater IA, TLA, and SLA scores than 886, although, these differences were not significant. This affirms previous rust bioassays that found most Arachis species, especially those in section Arachis, to be highly resistant to rust (Subrahmanyam et al., 1983; Fávero et al., 2009). Like this study, Pande and Rao (2001), Fávero et al. (2009), and Leal-Bertioli et al. (2015) found A. ipaënsis to be as susceptible or more susceptible than cultivated peanut. Fávero et al. (2009) also found sporulation on six accessions of A. stenosperma, two accessions of A. valida, and one accession of A. magna, which were not tested in this study. So far, high resistance to rust has not been identified in A. hypogaea germplasm (Power et al., 2019; Subrahmanyam et al., 1982; Pande and Rao, 2001; Fávero et al., 2009). The lack of high resistance to rust in cultivated germplasm was contributed in part by having a highly susceptible progenitor as well as the ploidy barrier between allotetraploid peanut and its diploid, highly resistant Arachis relatives.
This study builds upon previous reports by identifying rust resistant allotetraploids that are cross compatible to A. hypogaea and are therefore valuable to breeding programs, instead of focusing solely on diploid, wild Arachis species. A few studies have identified rust resistance QTLs derived from the A-genome species A. cardenasii GKP10017 and the B-genome species A. magna K 30097 (Khedikar et al., 2010; Sujay et al., 2012; Leal-Bertioli et al., 2015). Sujay et al. (2012) identified five rust resistance QTLs in the same linkage group that explained up to 63% to 83% phenotypic variation; these QTLs likely originated from A. cardenasii GKP 10017. The populations used in Sujay et al. (2012) both have A. hypogaea GPBD 4 as a parent, which has A. hypogaea ‘ICGV 86855] as a parent, which in turn originated from a cross between A. hypogaea and A. cardenasii GKP 10017 (Shirasawa et al., 2018). The QTL that explained the most rust phenotypic variation at 83% has been validated and introgressed into three cultivated varieties through marker-assisted backcrossing, improving yield by 56 to 96% in rust infected environments (Varshney et al., 2014). Leal-Bertioli et al. (2015) identified 13 rust resistance QTLs from A. magna K 30097. One of these QTLs contributed to four components of rust resistance, including IA, TLA, SLA, and incubation period, and explained up to 59% of phenotypic variation. Another resistance QTL explaining up to 35% of phenotypic variation was located in the same linkage group, just 25.4 to 33.1 cM away from the major QTL. These QTL identified by Leal-Bertioli et al. (2015) are distinct from those identified by Sujay et al. (2012) and can be pyramided into the same peanut cultivars to yield more effective and more robust resistance. All nine rust resistant allotetraploids identified in this study have unique Arachis parents compared to these two previous studies; therefore, any of these allotetraploids could be used to map new rust resistance QTL for further stacking of resistance QTLs in cultivated peanut. However, the greatest chance of identifying a major QTL would come from a mapping population using an allotetraploid that has a unique A- and B-genome species. For example, BatCor, BatDur1, BatDur2, BatSten1, GregSten, MagDio, and ValSten1 would be good candidates for future rust resistance QTL mapping.
One perceived limitation of this study was that both experiments were confined to in vitro bioassays using excised peanut leaves rather than having these in vitro experiments in addition to field evaluations. However, past rust field evaluations have been complicated by other fungal pathogens such as Colletotrichum spp., Leptosphaerulina crassiasca Sechet., early leaf spot, Myrothecium roridum Tode ex. Fr., and late leaf spot (Subrahmanyam et al., 1983; Sujay et al., 2012). Furthermore, while peanut rust is common in countries with warm, tropical climates, it only threatens U.S. peanut production every few years when brought by tropical storms (Bromfield, 1971; Subrahmanyam et al., 1985). When achieved, rust pressure is very inconsistent and fragmented within the same field, and results identifying resistant germplasm could easily result from avoidance of the pathogen instead of actual resistance. Lastly, the morphological differences, i.e., canopy structure, between wild Arachis species, allotetraploids, and cultivated peanut can add variations to pathogen pressure in the field. Despite these complications, Subrahmanyam et al. (1983) tested wild Arachis species and cultivated peanut germplasm in the field and in vitro and while complications in the field study from three other fungal pathogens were encountered, the reactions to rust were the same in the field and the in vitro experiments. Six of the wild Arachis accessions tested by Subrahmanyam et al. (1983) were also tested in this study and all were found to be highly resistant presenting no rust symptoms in both studies. Therefore, in vitro bioassays like this study have produced consistent results with previous studies, have similar results to field rust experiments, and have been used to map rust resistance QTL, indicating in vitro bioassays are sufficient for identifying rust resistant germplasm (Subrahmanyam et al., 1983; Khedikar et al., 2010; Sujay et al., 2012; Leal-Bertioli et al., 2015).
Conclusions
This study built upon previous reports by testing rust resistance in numerous synthetic allotetraploids and diploid wild Arachis species. Nine allotetraploids demonstrated high levels of resistance to rust, making them a better source of rust resistance than cultivated peanut germplasm, which has only been found to have moderate levels of resistance. These allotetraploids are cross-compatible with peanut cultivars and thus, available as a genetic resource for peanut breeders. A few of these unique allotetraploids will be used to map rust resistance QTLs so that they can be introgressed into peanut cultivars along with the previously identified rust resistance QTLs from A. cardenasii GKP 10017 and A. magna K 30097. The long-term goal of this study is to create rust resistant peanut cultivars that can protect yields in the U.S. and to increase yields in tropical, developing countries for farmers that cannot afford, or do not have access, to costly fungicides.
Acknowledgements
This work was supported by the Agriculture and Food Research Initiative Competitive Grant no. 2018-67013-28139, co-funded by the USDA National Institute of Food and Agriculture and the National Peanut Board, by the National Science Foundation (grant # MCB-1543922), and by the AFRI NIFA Fellowships Grant Program: Predoctoral Fellowships project accession no. 1019105 from the USDA National Institute of Food and Agriculture. The authors would like to express their appreciation to Micah Levinson of the Department of Horticulture, University of Georgia, Georgia, for assisting in data collection.
Literature Cited
Bertioli, D., Gao, D., Ballen-Taborda, C., Chu, Y., Ozias-Akins, P., Jackson, S.A., Holbrook, C.C., and Leal-Bertioli. S.C.M. 2021. Registration of GA-BatSten1 and GA-MagSten1, two induced allotetraploids derived from peanut wild relatives with superior resistance to leaf spots, rust and root-knot nematode. J. Plant Regist. 15: 372– 378.
Branch, W.D. 1996. Registration of ’Georgia Green’ peanut. Crop Sci. 36: 806.
Bromfield, K.R. 1971. Peanut rust: A review of literature. Proc. Amer. Peanut Res. Ed. Soc. 3: 111– 121.
Chu, Y., Stalker, H.T., Marasigan, K., Levinson, C.M., Gao, D., Bertioli, D.J., Leal-Bertioli, S.C.M., Holbrook, C.C., Jackson, S.A., and Ozias-Akins. P. 2021. Registration of three peanut allotetraploid interspecific hybrids resistant to leaf spot diseases. J. Plant Regist.
Clevenger, J.P., Korani, W., Ozias-Akins, P., and Jackson. S.A. 2018. Haplotype-based genotyping in polyploids. Front. Plant Sci. 9: 564.
de Mendiburu, F. 2021. Agricolae: Statistical Procedures for Agricultural Research. R package version 1.3-5. https://CRAN.R-project.org/package=agricolae
Fávero, A.P., Moraes, S.A., Garcia, A.A.F., Valls, J.F.M., and Vello. N.A. 2009. Characterization of rust, early and late leaf spot resistance in wild and cultivated peanut germplasm. Sci. Agric. 66: 110– 117.
Gowda, M.V.C., Motagi, B.N., Naidu, G.K., Diddimani, S.B., and Sheshagiri. R. 2002. GPBD 4: A Spanish bunch groundnut genotype resistant to rust and late leaf spot. Int. Arachis Newsl. 22: 29– 32.
Guimaraes, L.A., Pereira, B.M., Araujo, A.C.G., Guimaraes, P.M., and Brasileiro. A.C.M. 2017. Ex vitro hairy root induction in detached peanut leaves for plant–nematode interaction studies. Plant Methods. 13: 25.
Khedikar, Y.P., Gowda, M.V.C., Sarvamangala, C., Patgar, K.V., Upadhyaya, H.D., and Varshney. R.K. 2010. A QTL study on late leaf spot and rust revealed one major QTL for molecular breeding for rust resistance in groundnut (Arachis hypogaea L.). Theor. Appl. Genet. 121: 971– 984.
Korani, W., Clevenger, J.P., Chu, Y., and Ozias-Akins. P. 2019. Machine learning as an effective method for identifying true single nucleotide polymorphisms in polyploid plants. Plant Genome. 12: 180023.
Leal-Bertioli, S.C.M., Cavalcante, U., Gouveia, E.G., Ballen-Taborda, C., Shirasawa, K., Guimaraes, P.M., Jackson, S.A., Bertioli, D.J., and Moretzsohn. M.C. 2015. Identification of QTLs for rust resistance in the peanut wild species Arachis magna and the development of KASP markers for marker assisted selection. Genes/Genomes/Genet. 5: 1403– 1413.
Levinson, C.M., Marasigan, K.M., Chu, Y., Stalker, H.T., Holbrook, C.C., Ni, X., Williams, W.P, and Ozias-Akins. P. 2020. Resistance to fall armyworm (Lepidoptera: Noctuidae) feeding identified in nascent allotetraploids cross-compatible to cultivated peanut (Arachis hypogaea L.). Peanut Sci. 47: 123– 134.
Pande, S., and Rao. J.N. 2001. Resistance of wild Arachis species to late leaf spot and rust in greenhouse trials. Plant Dis. 85: 851– 855.
Power, I.L. 2014. Characterizing peanut rust resistance: Determining its mechanisms, and the genetics of the peanut host and Puccinia arachidis (Doctoral dissertation). University of Georgia, Athens, GA.
Power, I.L. Tillman, B.L., Brenneman, T.B., Kemerait, R.C., Stevenson, K.L., and Culbreath. A.K. 2019. Field evaluations and components of peanut rust resistance in newly developed breeding lines. Peanut Sci. 46: 22– 36.
R Core Team. 2021. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Savary, S., Rao, P.V.S., and Zadoksa. J.C. 1989. Scale of reaction types of groundnut to Puccinia arachidis Speg. J. Phytopathol. 124: 259– 266.
Shirasawa, K., Bhat, R.S., Khedikar, Y.P., Sujay, V., Kolekar, R.M., Yeri, S.B., Sukruth, M., Cholin, S., Asha, B., Pandey, M.K., Varshney, R.K., and Gowda. M.V.C 2018. Sequencing analysis of genetic loci for resistance for late leaf spot and rust in peanut (Arachis hypogaea L.). Front. Plant Sci. 9: 1727.
Smith, N.B. and Littrell. R.H. 1980. Management of peanut foliar diseases with fungicides. Plant Dis. 64: 356– 360.
Subrahmanyam, P. 1997. Rust. In: Kokalis-Burelle, N., Porter, D.M., Rodriguez-Kabana, R., Smith, D.H. and Subrahmanyam P. (Eds.), Compendium of Peanut Diseases. Amer. Phytopath. Soc., St. Paul, MN, pp. 31– 33.
Subrahmanyam, P., McDonald, D., Gibbons, R.W., Nigam, S.N., and Nevill. D.J. 1982. Resistance to rust and late leafspot diseases in some genotypes of Arachis hypogaea. Peanut Sci. 9: 6– 10.
Subrahmanyam, P., McDonald, D., Gibbons, R.W., and Reddy. L.J. 1985. Peanut rust: A major threat to peanut production in the semiarid tropics. Plant Dis. 69: 813– 819.
Subrahmanyam, P., Moss, J.P, and Rao. V.R. 1983. Resistance to peanut rust in wild Arachis species. Plant Dis. 67: 209– 212.
Subrahmanyam, P., Williams, J.H., McDonald, D., and Gibbons. R.W. 1984. The influence of foliar diseases and their control by selective fungicides on a range of groundnut (Arachis hypogaea L.) genotypes. Ann. Appl. Biol. 104: 467– 476.
Sujay, V., Gowda, M.V.C., Pandey, M.K., Bhat, R.S., Khedikar, Y.P., Nadaf, H.L., Gautami, B., Sarvamangala, C., Lingaraju, S., Radhakrishan, T., Knapp, S.J., and Varshney. R.K. 2012. Quantitative trait locus analysis and construction of consensus genetic map for foliar disease resistance based on two recombinant inbred line populations in cultivated groundnut (Arachis hypogaea L.). Mol. Breed. 30: 773– 788.
Varshney, R.K., Pandey, M.K., Janila, P., Nigam, S.N., Sudini, H., Gowda, M.V.C, Sriswathi, M., Radhakrishnan, T., Manohar, S.S., and Nagesh. P. 2014. Marker-assisted introgression of a QTL region to improve rust resistance in three elite and popular varieties of peanut (Arachis hypogaea L.). Theor. Appl. Genet. 127: 1771– 1781.
Williams-Woodward, J. 2013. 2011: Georgia plant disease loss estimates. The University of Georgia Cooperative Extension Service Annual Publication, Athens, GA, pp. 102– 104.
Notes
- Institute of Plant Breeding, Genetics and Genomics, University of Georgia, Athens, GA, U.S.. [^]
- Plant Pathology Department, University of Georgia, Athens, GA, U.S.. [^]
- Horticulture Department, University of Georgia, Tifton, GA, U.S.. [^]
- Plant Pathology Department, University of Georgia, Tifton, GA, U.S.. [^]
- Crop and Soil Sciences, North Carolina State University, Raleigh, NC, U.S.. [^]
- Small Grains and Potato Germplasm Research Unit, USDA-ARS, Aberdeen, ID, U.S.. [^] *Corresponding author Email: pozias@uga.edu
Author Affiliations