Introduction
Cultivated peanut (Arachis hypogaea L.) is an allotetraploid derived from a single recent hybridization event between A. duranensis and A. ipaensis (Kochert et al., 1996; Moretzsohn et al., 2013; Bertioli et al., 2015, 2019). Relative to some domesticated crops such as the common bean, Phaseolus vulgaris (Zizumbo-Villarreal et al., 2005), peanut cultigens possess little genetic variation (Kochert et al., 1991; Ferguson et al., 2004), even those collected from peanut’s center of origin in South America (Halward et al., 1991; Moretzsohn et al., 2004). In contrast to the limited resistance to pests and diseases available within the germplasm pool for Arachis hypogaea (Stalker, 2017), many wild Arachis species possess high levels of genetic diversity, and importantly, resistance to various biotic stressors (Stalker et al., 2016; Stalker, 2017). Accessions from at least 30 species within Section Arachis, the secondary gene pool for cultivated peanut (Krapovickas and Gregory, 2007; Smýkal et al., 2015), have demonstrated resistance to diseases and insects (Stalker, 2017). The U.S. National Plant Germplasm System currently has approximately 500 available wild species accessions, approximately 200 of which are from Section Arachis.
Despite the potential of wild species for improving cultivated peanut, most cultivars in the U.S. do not have wild Arachis in their ancestries (Stalker, 2017). Nonetheless, the few examples descended from wild species have had significant impact. All nematode resistance in U.S. cultivars—i.e., COAN (Simpson and Starr, 2001), NemaTAM (Simpson et al., 2003), Tifguard (Holbrook et al., 2008), Webb (Simpson et al., 2013), Georgia-14N (Branch and Brenneman, 2015), and TifNV-High O/L (Holbrook et al., 2017)—is derived from TxAG-6, a complex interspecific hybrid generated from A. cardenasii, A. diogoi (formerly A. chacoense), and A. batizocoi (Simpson et al., 1993). Arachis cardenasii is also in the heritage of Bailey (Isleib et al., 2011), the popular Virginia cultivar with some resistance to early and late leaf spots, Sclerotinia blight, Cylindrocladium black rot, tomato spotted wilt virus, and Athelia rolfsii (Curzi) C.C. Tu & Kimbr. GP-NC WS 13, a North Carolina State University (NCSU) germplasm release generated from A. hypogaea and A. cardenasii (Stalker et al., 2002), is two breeding cycles removed from Bailey. The International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) has also released several cultivars developed from interspecific crosses containing A. batizocoi, A. cardenasii, A. duranensis, A. stenosperma, and A. villosa with resistances to peanut rust, late leaf spot, and viruses (Gowda et al., 2002; Singh et al., 2003; Stalker, 2017).
Multiple studies have evaluated wild Arachis species for resistance to early and late leaf spots, rust, viruses, and nematodes (Subrahmanyam et al., 1983b; a, 1985, 2001; Nelson et al., 1989; Holbrook and Noe, 1990; Reddy et al., 2000; Pande and Rao, 2001; Sharma et al., 2017; Stalker, 2017). However, considerably less attention has been paid to soilborne disease resistance. An expansive evaluation by Tallury et al. (2013) evaluated 110 accessions from 23 Arachis species for resistance to Cylindrocladium black rot (CBR) and Sclerotinia blight. Considerable variation in resistance was found for CBR, but only A. glandulifera exhibited significantly more resistance than A. hypogaea for Sclerotinia blight (Tallury et al., 2013). Pande et al. (1994) included one each of A. chacoense (= diogoi, PI 276235), monticola (PI 497260), A. stenosperma (PI 497579), and an interspecific hybrid of A. hypogaea and A. cardenasii in a greenhouse resistance assay for At. rolfsii. Bera et al. (2016) at the Indian Council of Agricultural Research Directorate of Groundnut Research (ICAR-DGR) evaluated a total of 25 accessions of 11 species from four sections of Arachis for resistance to At. rolfsii. The most resistant accessions, A. pusilla DGR 12047 and A. appressipila ICG 8945, had 13 and 14% mortality, respectively. Researchers at ICAR-DGR also developed NRCG CS85, a multiple disease-resistant genotype derived from A. kretschmeri, to create a mapping population (Dodia et al., 2019) and to investigate mechanisms of resistance to southern blight (Bosamia et al., 2020).
The objective of this study was to evaluate a small subset of accessions within the Section Arachis for resistance to At. rolfsii in the greenhouse. Formerly known as Sclerotium rolfsii (Xu et al., 2010), this cosmopolitan pathogen is the most damaging soilborne disease of peanut in the U.S. (Backman and Brenneman, 1997). In Georgia alone, an estimated $91.4M was lost in yield and control costs from At. rolfsii in 2017 (Little, 2017). Disease-resistant peanut cultivars developed from resistant wild species would save growers millions of dollars and mitigate health impacts by reducing fungicide applications (Fisher et al., 2018).
Materials and Methods
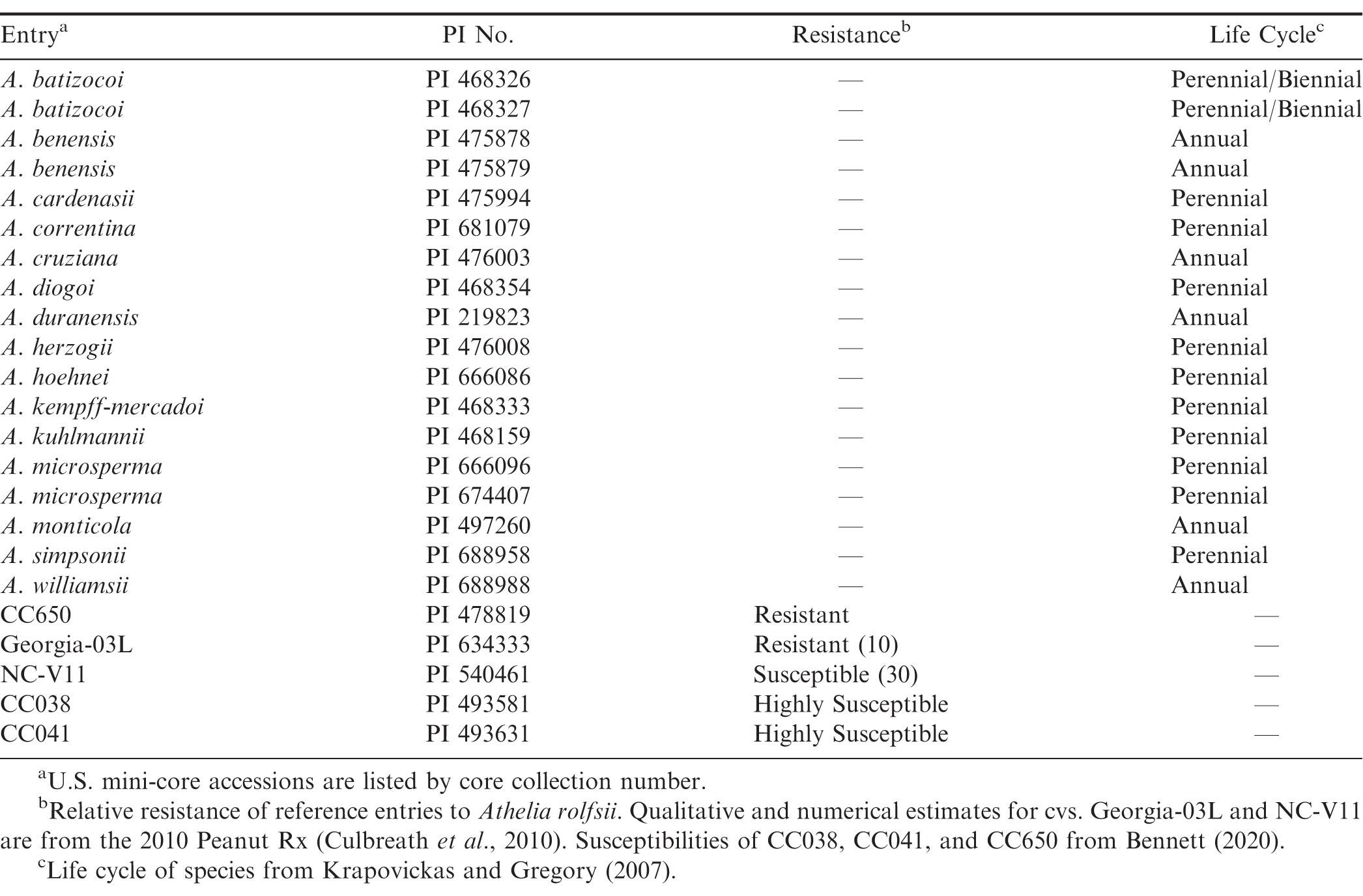
A total of 18 PIs from the U.S. National Plant Germplasm System representing 15 section Arachis species (batizocoi, benensis, cardenasii, correntina, cruziana, diogoi, duranensis, herzogii, hoehnei, kempff-mercadoi, kuhlmannii, microsperma, monticola, simpsonii, williamsii) were screened in the greenhouse for resistance to At. rolfsii (Table 1). Resistant and susceptible reference genotypes, the runner cv. Georgia-03L (Branch, 2004), U.S. mini-core accessions CC650 (PI 478819), CC038 (PI 493581), and CC041 (PI 493631)(Bennett and Chamberlin, 2020) were included to compare the greenhouse system to a previous-used growth chamber system (Bennett, 2020). The susceptible Virginia cv. NC-V11 (Wynne et al., 1991), rated “30” for white mold resistance by Peanut Rx (Culbreath et al., 2010) was also included.
Plant Preparation.
Because of limited seed and inconsistent germination of most wild Arachis accessions, they were vegetatively propagated from May to September 2019 to obtain plants for the experiments. Mother plants were maintained in pots (33-cm-top diam., 20-cm-bottom diam., 15-cm tall) filled with the soilless mix BM7-35 (Berger). Greenhouse temperatures were set at 22 to 32 C. A small amount of Rhizobium inoculant (Guard-N; Verdesian Life Sciences) was applied by dipping the seed into the inoculant powder immediately before planting or by placing ca. 3 mm3 of the inoculant into the planting hole.
Cuttings were taken from branches with 3 to 4 nodes, and leaves, pegs, and flowers were removed from the lowermost 1 to 2 nodes. The bottom nodes were dipped in rooting powder (Garden Safe TakeRoot; Spectrum Brands Holdings) before placing in 9.5-diam. pots filled with a soilless propagation mix (Sunshine Redi-Earth Plug and Seedling; Sungro). Rhizobium inoculant was applied to the planting hole as previously described before cuttings were inserted into the potting mix. Potted cuttings were placed in an 0.9-m-wide x 3.3-m-long x 0.9-m-high enclosed humidity chamber constructed from 2.54-diam. polyvinyl chloride piping and 6-mil Solar-Ice polyethylene film (now Luminance, RPC BPI Agriculture). The humidity chamber was partially covered with 50% Aluminet (Ecologic Technologies, Inc.) to further reduce heat from infrared radiation. Overhead misting lines (Orbit Irrigation Products) provided high humidity by misting approximately 5 sec every 5 min. After four weeks, cuttings were monitored weekly for root development by gently pulling on the stem. Rooted cuttings were removed from the chamber and placed in shade for 2-3 weeks to acclimate to lower humidity. Cuttings were then transplanted into 15-cm-diam. pots filled with BM7-35 soilless mix and moved into full light on greenhouse benches. A 46-cm-long x 6.35-diam. acrylic rod (McMaster-Carr) was inserted into each pot, and top-heavy and trailing plants were tied to the rod with paper wire twist ties. In order to prolong the life of biennial and annual species (Kvien and Ozias-Akins, 1991), the vegetatively propagated plants were monitored weekly to remove any pegs that had developed. Plants were also pruned as needed to keep branches from extending ca. 30 cm beyond the top of the acrylic rod.
For the resistant and susceptible A. hypogaea reference genotypes, three seeds were planted with Rhizobium inoculant in 15-cm-diam pots filled with the BM7-35 potting mix. Pots were planted eight weeks before inoculation and thinned to one plant after emergence. All wild and cultivated plants were fertilized with 15 mL of NPK 14-14-14 slow-release product (Osmocote Smart Release Flower and Vegetable; ScottsMiracle-Gro). In addition, micronutrients were applied by adding Fertileader Vital (300 mL/379 L water; Timac Agro USA) to the irrigation water until a minimum of 2 weeks before inoculation.
Athelia rolfsii Inoculations.
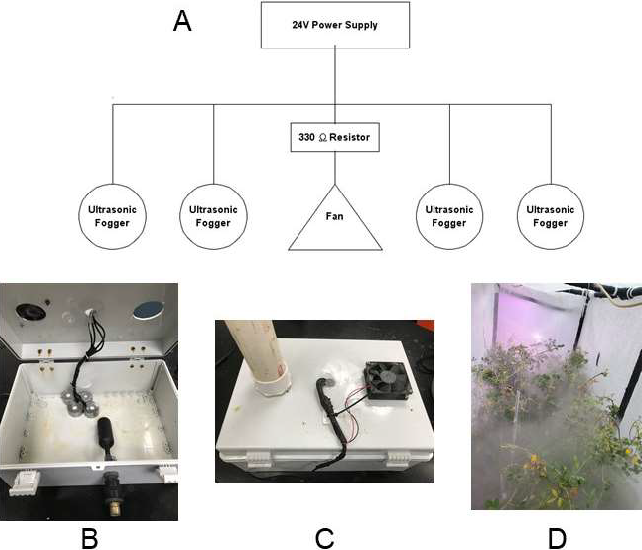
Athelia rolfsii mycelial plugs were prepared and plant inoculations were conducted as described in a previous study (Bennett, 2020) with the exception that plants were inoculated on the main stem, usually at the 2nd node, approximately 30-60 mm from the soil line. Because A. microsperma PI 674407 had a particularly bushy growth habit, the impeding branches were tied upwards with string to facilitate inoculation and data collection. Plants were placed inside the humidity chamber, where high humidity was maintained for part of the day with an ultrasonic transducer fogging system on each end of the chamber. The fogging system, hooked up to a water hose, was constructed using a 39 cm x 30 cm x 17 cm project box, a water tank float to maintain proper water level, a 24 V computer fan, four 24 V ultrasonic transducers, and a 24 V power supply (Figure 1). Each ultrasonic transducer was able to diffuse between 200-400 mL of water per hour. The fogging systems operated 9 hr per day, with one running between 0600 and 1500 hr, and the other between 1100 and 2000 hr. Temperature and relative humidity were monitored each hr with three HOBO U23 data loggers (Onset Computer Corp.) starting 25 Feb. 2020. Main stem lesion length was measured 4, 6, 10, and 12 d after inoculation using a digital caliper (Mitutoyo America). Because mycelium frequently obscured lesions, it is often faster to measure mycelium than to measure lesions. Thus, mycelium measurements were taken to see if they could be substituted for lesion measurements. If the plant died before the end of the experiment, measurements for remaining days were recorded as missing data. The experiment (trial) was conducted 11 times between 9 Feb. 2020 to 29 Apr. 2020.
Data Analyses.
Plant pots were arranged inside the chamber in a randomized complete block design with two replications. Data were analyzed using SAS Version 9.4 (SAS Institute). Differences among entries in lesion length and mycelial growth were determined using repeated measures ANOVA in PROC MIXED with TOEP covariance structure. Trial and block(trial) were used as random variables in the model. Differences among and within entries at 4, 6, 10, and 12 d after inoculation were analyzed using the SLICE option. Correlation analysis between lesion and mycelium lengths were conducted using PROC CORR. Area under the disease progress curves (AUDPCs; Shaner and Finney, 1977) for lesion length were estimated and analysed using PROC GLIMMIX. AUDPC means were compared using a split-plot design with trial as the whole plot and entry as the subplot, and the SLICE option was used to examine differences among trials and entries. All pairwise comparisons were adjusted for Type I error with the ADJUST = TUKEY option at α = 0.05.
Results and Discussion
Some Arachis species were easier to propagate vegetatively than others, so there were fewer plants for some species, e.g. A. correntina, A batizocoi PI 468326, and A. cruziana (Table 2). During the experiments, the temperature inside the humidity chamber varied from 20 to 37 C but reached maximum temperatures of 27 C or greater on 90% of the days recorded by the data loggers. Temperatures between 27 and 30 C are considered to be optimal for At. rolfsii growth (Punja, 1985). In addition, higher temperatures were observed in April than in March and February. Median and maximum temperatures in April were 0.9 C and 6.8 C greater, respectively, than those observed in February. On 64% of the logged data times, relative humidity inside the chamber was ≥ 90%, and daily maximum relative humidity reached 98% or greater on 61 of the 63 logged days.
Lesion Length and Mycelial Growth.
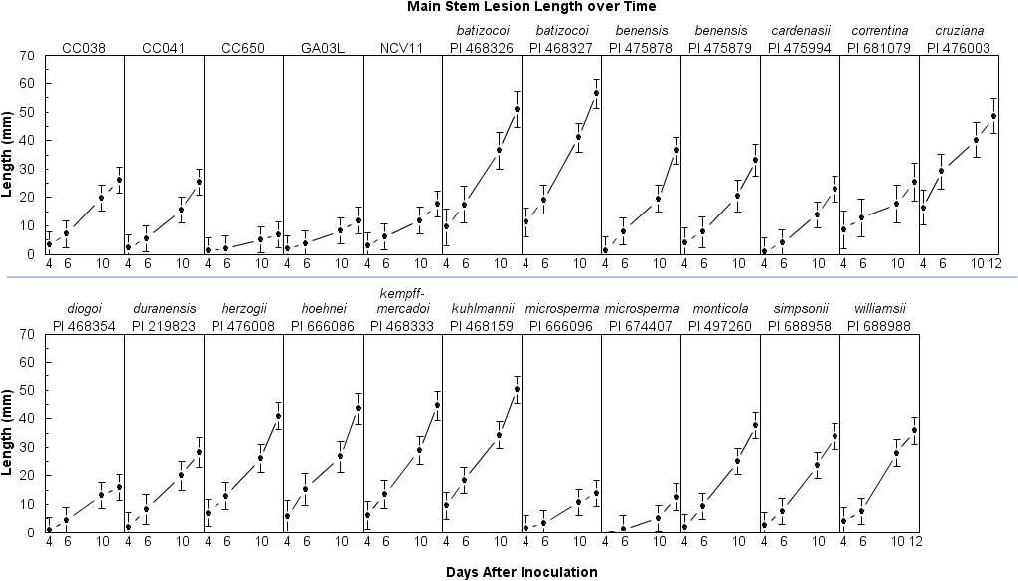
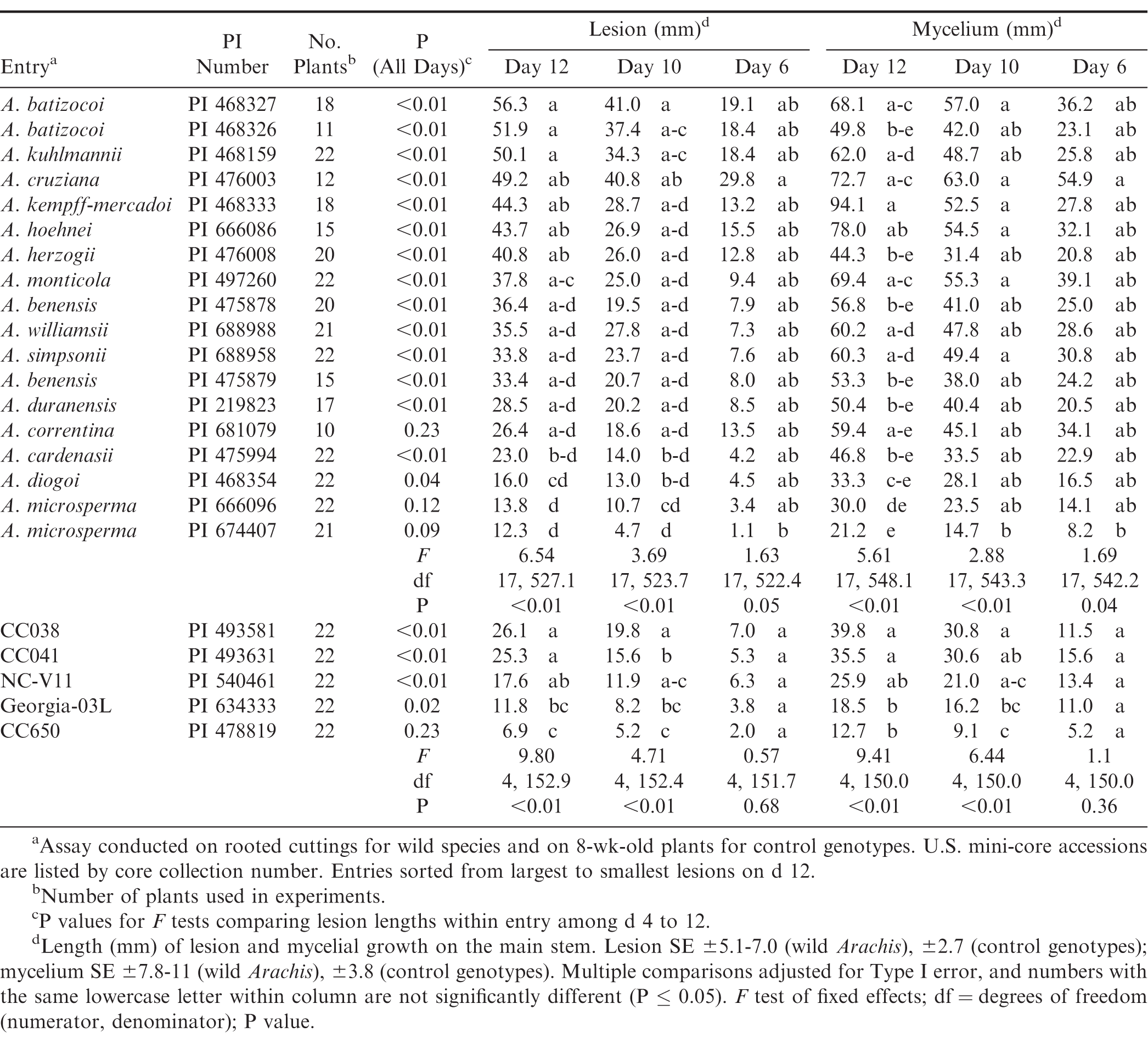
In the repeated measures analyses of lesion length, the interaction between entry and time was significant, indicating that differences among entries depended on the day of observation (F = 1.44; df = 51, 827; P = 0.03). Lesion lengths among entries differed on d 6, 10, and 12 (Table 2) but not on d 4 (F = 0.63; df = 22, 693.1; P = 0.91). On d 12, the longest lesions (> 50 mm) were found on the two A. batizocoi accessions, and A. kuhlmannii (Table 2). The shortest lesions (≤ 23 mm) were found on A. cardenasii, A diogoi, and A. microsperma PI 674407 and PI 666096. When sliced by entry, lesions lengths were significantly different over time for all entries except A. correntina, and the two A. microsperma PIs (Table 2).
The interaction between entry and time (F = 1.41; df = 51, 834; P < 0.03) was also significant for mycelial growth. Significant differences among entries were found for all days except d 4 (F = 0.53; df = 17, 541.3; P = 0.94). Mycelial growth occasionally decreased on some plants between d 10 and 12, but entries with the most mycelium (> 52 mm) for both days were A. kempff-mercadoi, A. hoehnei, A cruziana, A. monticola, and A. batizocoi PI 468327 (Table 2). These entries sustained significantly more mycelial growth than both A. microsperma entries by d 12. The correlation between mycelial growth and lesion length over time was considerable (r = 0.74; P < 0.01), but not as strong as in a previous study consisting only of A. hypogaea entries (r = 0.92; P < 0.01; Bennett, 2020). When the relationship was examined by day, the best correlation was found on d 10 (r = 0.76; P < 0.01), followed by d 12 (r = 0.73; P < 0.01), d 6 (r = 0.52; P < 0.01), and d 4 (r = 0.36; P < 0.01). Most entries had strong correlations (r > 0.70) between mycelial growth and lesion length except A. monticola (r = 0.40; P < 0.01), A. cruziana (r = 0.61; P < 0.01), A. diogoi (r = 0.62; P < 0.01), and A. kempff-mercadoi (r = 0.69; P < 0.01). Interestingly, A. monticola supported substantial mycelial growth but had comparatively smaller lesions on d 10 and 12. Thus, it appears that mycelial growth may not be a suitable indicator of resistance for comparisons among Arachis species.
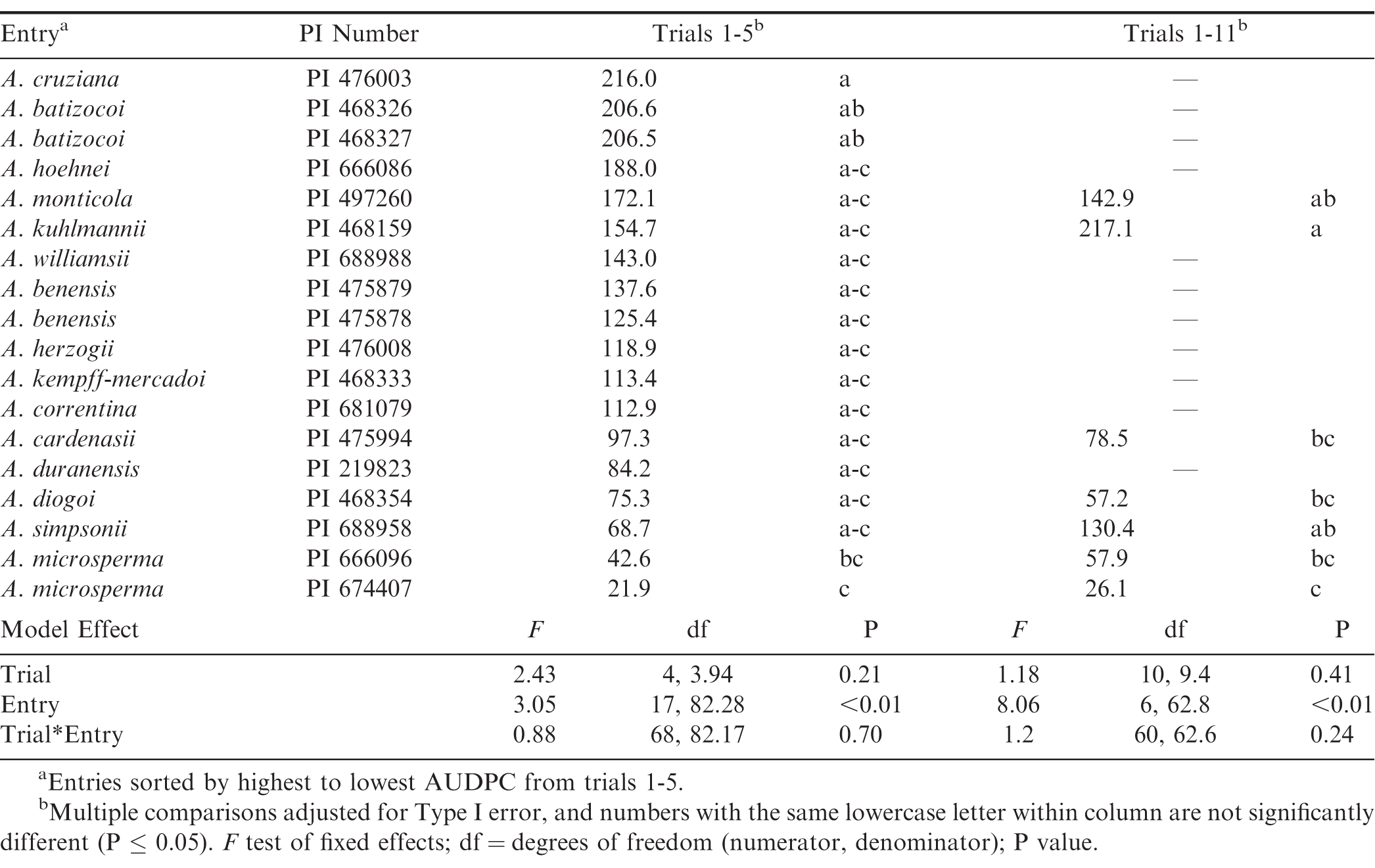
Area Under the Disease Progress Curve (AUDPC).
Area under the disease progress curves were also examined. Missing data for the 11 entries with fewer plants resulted in non-convergence of the model, so data from all entries in trials 1 to 5 were used in one analysis, and data from the seven entries used in all trials were analyzed separately (Table 3). For both analyses, the effects of trial (trials 1-5, P = 0.21; trials 1-11, P = 0.41) and trial x entry (P > 0.24) were not significant. However, the effect of entry was significant (P < 0.01). In the first five trials, the highest AUDPC was in A cruziana, followed by the two A. batizocoi accessions; the two A. microsperma accessions had the lowest AUDPCs. When the seven entries were compared over all trials, A. kuhlmannii had the highest mean AUDPC, which was significantly greater than those of A. cardenasii, A. diogoi, and both A. microsperma PIs. To note, A. simpsonii was among the more resistant entries in trials 1-5 but appeared considerably more susceptible in the analyses of all trials. Similarly, the AUDPC of A. kuhlmannii increased by 62 mm when the later trials were added. These changes in susceptibility may be due to the higher temperatures observed in the later trials, and A. simpsonii susceptibility to At. rolfsii may be particularly temperature sensitive. Pande et al. (1994) also observed that plant mortality to At. rolfsii increased with higher temperatures in some genotypes.
Compared to a previous study (Bennett, 2020), the greenhouse humidity chamber used here resulted in less severe disease, especially among the susceptible control genotypes CC038 and CC041. The difference in results is likely due to the constant optimum temperature maintained by growth chambers in that study. However, CC038 and CC041 were significantly more susceptible to A. rolfsii than the resistant genotypes Georgia-03L and CC650 (Table 2), consistent with recent laboratory and field studies (Bennett, 2020; Bennett and Chamberlin, 2020). The susceptible cultivar NC-V11 was numerically intermediate in lesion length and mycelial growth to the susceptible and resistant genotypes and differed statistically only from CC650 in lesion length on d 12. Both the greenhouse and growth chamber assays were better at identifying highly susceptible and some highly resistant genotypes than discriminating among intermediate entries (Bennett, 2020). Despite this limitation, laboratory assays are less likely to be influenced by canopy microclimates than field evaluations because high levels of humidity are easier to maintain. Arachis spp. vary considerably in canopy architecture, e.g. ranging from the relatively short and ramose A. microsperma to the little-branched, 1-m-tall mainstem (and up to 4-m-long lateral branches) of A. batizocoi (Krapovickas and Gregory, 2007).
Comparisons between the reference A. hypogaea genotypes and wild species were not made due to differences in plant age at inoculation. Several studies have observed that susceptibility to At. rolfsii decreases with plant age (Pande et al., 1994; Pratt and Rowe, 2002; Bekriwala et al., 2016), yet the younger A. hypogaea entries used in this study had smaller lesions than most of the wild species. Additional work is needed to determine if vegetative propagation enhanced disease susceptibility. In future studies, an ordinal rating ranking, such as the Florida scale for leaf spot (Chiteka et al., 1988), will added to accommodate informative qualitative characters such as wilting and death. Nonetheless, there were significant differences among the wild Arachis accessions in susceptibility to At. rolfsii as indicated by lesion length, mycelial growth, and AUDPCs. Within the wild species, A. microsperma PI 674407 and PI 666096 exhibited small lesions, mycelial growth, and AUDPC. Both accessions have previously shown resistance to Cylindrocladium black rot (Tallury et al., 2013). Arachis cardenasii PI 475994 and A. diogoi PI 468354 also had relatively small lesions. PI 475994 has high resistance to Meloidogyne javanica race 3 (Sharma et al., 2002), and PI 468354 is resistant to tomato spotted wilt virus (Lyerly et al., 2002; Wang et al., 2009). Since the three species have an A genome, facilitating introgression into cultivated peanut, these four accessions may merit additional evaluation as candidates for pre-breeding.
Acknowledgements
The authors thank anonymous reviewers for providing helpful comments and the following Oklahoma State University undergraduate students for their help with this project: Destiny Burrell, Barrett Cosby, Ivy Hover, Paula Lor, and Peter Vang. This research was supported by USDA-ARS CRIS Project No. 3072-21220-008-00D. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Literature Cited
Backman, P.A., and Brenneman. T.B. 1997. Stem rot. Pages 36– 37. In Compendium of Peanut Diseases. American Phytopathological Society, St. Paul, Minnesota.
Bekriwala, T.H., Nath, K. and Chaudhary. D.A. 2016. Effect of age on susceptibility of groundnut plants to Sclerotium rolfsii Sacc. caused stem rot disease. J. Plant Pathol. Microbiol. 7: 386.
Bennett, R.S. 2020. Growth chamber assay for evaluating resistance to Athelia rolfsii. Peanut Sci. 47: 25– 32.
Bennett, R.S., and Chamberlin. K.D. 2020. Resistance to Athelia rolfsii and web blotch in the U.S. mini-core collection. Peanut Sci. 47: 17– 24.
Bera, S.K., Kamdar, J.H. Kasundra, S.V. and Thirumalaisami. P.P. 2016. Identification of groundnut genotypes and wild species resistant to stem rot using an efficient field screening technique. Electron. J. Plant Breed. 7: 61– 70.
Bertioli, D.J., Cannon, S.B. Froenicke, L. Huang, G. Farmer, A.D. et al. 2015. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 47: 438.
Bertioli, D.J., Jenkins, J. Clevenger, J. Dudchenko, O. Gao, D. et al. 2019. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 51: 877– 884.
Bosamia, T.C., Dodia, S.M. Mishra, G.P. Ahmad, S. Joshi, B. et al. 2020. Unraveling the mechanisms of resistance to Sclerotium rolfsii in peanut (Arachis hypogaea L.) using comparative RNA-Seq analysis of resistant and susceptible genotypes. PLoS ONE 15: e0236823.
Branch, W.D. 2004. Registration of ‘Georgia-03L’ peanut. Crop Sci. 44: 1485– 1486.
Branch, W.D., and Brenneman. T.B. 2015. Registration of ‘Georgia-14N’ Peanut. J. Plant Regist. 9: 159– 161.
Chiteka, Z.A., Gorbet, D.W. Shokes, F.M. Kucharek, T.A. and Knauft. D.A. 1988. Components of resistance to late leafspot in peanut. I. Levels and variability - implications for selection. Peanut Sci. 15: 25– 30.
Culbreath, A., Beasley, J. Kemerait, R. Prostko, E. Brenneman, T. et al. 2010. Peanut Rx: Minimizing diseases of peanut in the southeastern United States, the 2010 version of the peanut disease risk index. Univ. of Georgia Coop. Ext. Serv., Athens, GA.
Dodia, S.M., Joshi, B. Gangurde, S.S. Thirumalaisamy, P.P. Mishra, G.P. et al. 2019. Genotyping-by-sequencing based genetic mapping reveals large number of epistatic interactions for stem rot resistance in groundnut. Theor. Appl. Genet. 132: 1001– 1016.
Ferguson, M.E., Bramel, P.J. and Chandra. S. 2004. Gene diversity among botanical varieties in peanut (Arachis hypogaea L.). Crop Sci. 44: 1847– 1854.
Fisher, M.C., Hawkins, N.J. Sanglard, D. and Gurr. S.J. 2018. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 360: 739– 742.
Gowda, M.V.C., Motagi, B.N. Naidu, G.K. Diddimani, S.B. and Sheshagiri. R. 2002. GPBD 4: a Spanish bunch groundnut genotype resistant to rust and late leaf spot. Int. Arachis Newsl. 22: 29– 32.
Halward, T.M., Stalker, H.T. Larue, E.A. and Kochert. G. 1991. Genetic variation detectable with molecular markers among unadapted germ-plasm resources of cultivated peanut and related wild species. Genome 34: 1013– 1020.
Holbrook, C.C., and Noe. J.P. 1990. Resistance to Meloidogyne arenaria in Arachis spp. and the implications on development of resistant peanut cultivars. Peanut Sci. 17: 35– 38.
Holbrook, C.C., Timper, P. Culbreath, A.K. and Kvien. C.K. 2008. Registration of ‘Tifguard’ peanut. J. Plant Regist. 2: 92– 94.
Holbrook, C.C., Ozias-Akins, P. Chu, Y. Culbreath, A.K. Kvien, C.K. et al. 2017. Registration of ‘TifNV-High O/L’ peanut. J. Plant Regist. 11: 228– 230.
Isleib, T.G., Milla-Lewis, S.R. Pattee, H.E. Copeland, S.C. Zuleta, M.C. et al. 2011. Registration of ‘Bailey’ peanut. J. Plant Regist. 5: 27.
Kochert, G., Halward, T. Branch, W.D. and Simpson. C.E. 1991. RFLP variability in peanut (Arachis hypogaea L.) cultivars and wild species. Theor. Appl. Genet. 81: 565– 570.
Kochert, G., Stalker, H.T. Gimenes, M. Galgaro, L. and Moore. K. 1996. RFLP and cytogenetic evidence for the progenitor species of allotetraploid cultivated peanut, Arachis hypogaea (Leguminosae). Am. J. Bot. 83: 1282– 1291.
Krapovickas, A. & Gregory. W.C. 2007. Taxonomy of the Genus Arachis (Leguminosae). Translated by Williams D.E. & Simpson. C.E. Bonplandia 16 (Supl.):1-205. ISSN 0524-0476.
Kvien, C.K., and Ozias-Akins. P. 1991. Lack of monocarpic senescence in Florunner peanut. Peanut Sci. 18: 86– 90.
Little, E.L. 2017. 2017 Georgia Plant Disease Loss Estimates. Univ. of Georgia Coop. Ext. Serv., Annual Publication 102-10, Athens, GA.
Lyerly, J.H., Stalker, H.T. Moyer, J.W. and Hoffman. K. 2002. Evaluation of Arachis species for resistance to tomato spotted wilt virus. Peanut Sci. 29: 79– 84.
Moretzsohn, M.C., Gouvea, E.G. Inglis, P.W. Leal-Bertioli, S.C. Valls, J.F. et al. 2013. A study of the relationships of cultivated peanut (Arachis hypogaea) and its most closely related wild species using intron sequences and microsatellite markers. Ann. Bot. 111: 113– 126.
Moretzsohn, M., Hopkins, M.S. Mitchell, S.E. Kresovich, S. Valls, J.F.M. et al. 2004. Genetic diversity of peanut (Arachis hypogaea L.) and its wild relatives based on the analysis of hypervariable regions of the genome. BMC Plant Biol. 4: 11.
Nelson, S.C., Simpson, C.E. and Starr. J.L. 1989. Resistance to Meloidogyne arenaria in Arachis spp. germplasm. J. Nematol. 21 (4S): 654.
Pande, S., Narayana, R.J. Reddy, M.V. and McDonald. D. 1994. A technique to screen for resistance to stem rot caused by Sclerotium rolfsii in groundnut under greenhouse conditions. Indian J. Plant Prot. 22: 151– 158.
Pande, S., and Rao. J.N. 2001. Resistance of wild Arachis species to late leaf spot and rust in greenhouse trials. Plant Dis. 85: 851– 855.
Pratt, R.G., and Rowe. D.E. 2002. Enhanced resistance to Sclerotium rolfsii in populations of alfalfa selected for quantitative resistance to Sclerotinia trifoliorum. Phytopathology 92: 204– 209.
Punja, Z.K. 1985. The biology, ecology, and control of Sclerotium rolfsii. Annu. Rev. Phytopathol. 23: 97– 127.
Reddy, A.S., Reddy, L.J. Mallikarjuna, N. Abdurahman, M.D. Reddy, Y.V. et al. 2000. Identification of resistance to peanut bud necrosis virus (PBNV) in wild Arachis germplasm. Ann. Appl. Biol. 137: 135– 139.
Shaner, G., and Finney. R.E. 1977. The effect of nitrogen fertilization on the expression of slow-mildewing resistance in Knox wheat. Phytopathology 67: 1051– 1056.
Sharma, S., Pandey, M.K. Sudini, H.K. Upadhyaya, H.D. and Varshney. R.K. 2017. Harnessing genetic diversity of wild species for genetic enhancement of cultivated peanut. Crop Sci. 57: 1121– 1131.
Sharma, S.B., Reddy, L.J. Bramel, P.J. and Ansari. M.A. 2002. Resistance to Meloidogyne javanica race 3 in the Arachis gene pool. Nematol. Mediterr. 30: 221– 225.
Simpson, C.E., Nelson, S.C. Starr, J.L. Woodard, K.E. and Smith. O.D. 1993. Registration of TxAG-6 and TxAG-7 peanut germplasm lines. Crop Sci. 33: 1418– 1418.
Simpson, C.E., and Starr. J.L. 2001. Registration of ‘COAN’ Peanut. Crop Sci. 41: 918.
Simpson, C.E., Starr, J.L. Baring, M.R. Burow, M.D. Cason, J.M. et al. 2013. Registration of ‘Webb’ peanut. J. Plant Regist. 7: 265– 268.
Simpson, C.E., Starr, J.L. Church, G.T. Burow, M.D. and Paterson. A.H. 2003. Registration of “NemaTAM” peanut. Crop Sci. 43: 1561– 1562.
Singh, A.K., Dwivedi, S.L. Pande, S. Moss, J.P. Nigam, S.N. et al. 2003. Registration of rust and late leaf spot resistant peanut germplasm lines. Crop Sci. 43: 440– 441.
Smýkal, P., Coyne, C.J. Ambrose, M.J. Maxted, N. Schaefer, H. et al. 2015. Legume crops phylogeny and genetic diversity for science and breeding. Crit. Rev. Plant Sci. 34: 43– 104.
Stalker, H.T. 2017. Utilizing wild species for peanut improvement. Crop Sci. 57: 1102– 1120.
Stalker, H.T., Beute, M.K. Shew, B.B. and Isleib. T.G. 2002. Registration of five leaf spot-resistant peanut germplasm lines. Crop Sci. 42: 314– 316.
Stalker, H.T., Tallury, S.P. Seijo, G.R. and Leal-Bertioli. S.C. 2016. Biology, speciation, and utilization of peanut species. Pages 27– 66. In: Peanuts. Academic Press and Press, AOCS New York.
Subrahmanyam, P., Anaidu, R. Reddy, L.J. Kumar, P.L. and Ferguson. M.E. 2001. Resistance to groundnut rosette disease in wild Arachis species. Ann. Appl. Biol. 139: 45– 50.
Subrahmanyam, P., Ghanekar, A.M. Nolt, B.L. Reddy, D.V.R. and McDonald. D. 1983 a. Resistance to groundnut diseases in wild Arachis species. Pages 49– 55. In Proceedings of an International Workshop on Cytogenetics of Arachis. ICRISAT, Patancheru, India.
Subrahmanyam, P., Moss, J.P. McDonald, D. Rao, P.S. and Rao. V.R. 1985. Resistance to leaf spot caused by Cercosporidium personatum in wild Arachis species. Plant Dis. 69: 951– 954.
Subrahmanyam, P., Moss, J.P. and Rao. V.R. 1983 b. Resistance to peanut rust in wild Arachis species. Plant Dis. 67: 209– 212.
Tallury, S.P., Hollowell, J.E. Isleib, T.G. and Stalker. H.T. 2013. Greenhouse evaluation of Section Arachis wild species for Sclerotinia blight and Cylindrocladium black rot resistance. Peanut Sci. 41: 17– 24.
Wang, M.L., Pinnow, D.L. Barkley, N.A. and Pittman. R.N. 2009. Plant resistance to TSWV and seed accumulation of resveratrol within peanut germplasm and its wild relatives in the U.S. collection. Plant Pathol. J. 8: 53– 61.
Wynne, J.C., Coffelt, T.A. Mozingo, R.W. and Anderson. W.F. 1991. Registration of ‘NC-V11′ Peanut. Crop Sci. 31: 484– 485.
Xu, Z., Harrington, T.C. Gleason, M.L. and Batzer. J.C. 2010. Phylogenetic placement of plant pathogenic Sclerotium species among teleomorph genera. Mycologia 102: 337– 346.
Zizumbo-Villarreal, D., Colunga-GarcíaMarín, P. de la Cruz, E.P. Delgado-Valerio, P. and Gepts. P. 2005. Population structure and evolutionary dynamics of wild–weedy–domesticated complexes of common bean in a Mesoamerican region. Crop Sci. 45: 1073– 1083.
Notes
- First and second authors: Research Plant Pathologist and Biological Science Technician, USDA-ARS, Wheat, Peanuts and Other Field Crops Research Unit, Stillwater, OK 74075; Third author: Professor Emeritus, Department of Soil and Crop Sciences, Texas A&M Agrilife, Stephenville, TX 76401; Fourth author: Geneticist, USDA-ARS, Plant Genetics Resources Conservation Unit, Griffin, GA 30223; Fifth and sixth authors: Undergraduate Student and Professor, Department of Biosystems and Agricultural Engineering, Oklahoma State Univ., Stillwater, OK 74078; Seventh author: Assistant Professor, Department of Crop and Soil Sciences, North Carolina State University, Raleigh, NC 27695. [^] *Corresponding author Email: rebecca.bennett@usda.gov
Author Affiliations