Introduction
Prior to the discovery of high-oleic, traditional cultivated peanut (Arachis hypogaea L.) seed were typically found to have an oleic (O) to linoleic (L) fatty acid ratios from 1 to 2:1 (Branch et al., 1990). However, Norden et al., (1987) reported upon the first high-oleic fatty acid peanut genotype with an O/L ratio of 35 to 37:1. This particular genotype (F435) had a spanish bunch-type growth habit and small-seed size with the unusually High-O/L ratio. Subsequently, genetic inheritance studies found that the High-O/L ratio oil trait was controlled by two recessive genes, ol1 and ol2 (Moore and Knauft, 1989, Knauft et al., 1993, and Isleib et al., 1996). Numerous peanut cultivars have since been released primarily because the high-oleic trait extends the shelf-life of peanut and peanut products (Mozingo et al., 2004).
Branch (2000) released a Very High-O/L peanut cultivar, ‘Georgia Hi-O/L’ with an O/L ratio of 40:1. The Georgia Hi-O/L cultivar originated from a cross made in 1990 between two Georgia peanut breeding lines, GA-C330A x GA-T2636M. GA-T2636M is an induced mutant breeding line derived from exposing seed of ‘Georgia Runner’ (Branch, 1991) to 200 Gy (20 kRad) of gamma irradiation from a cobalt-60 source. Since then other sources of high-oleic peanut mutants have been induced by gamma-rays (Nadaf et al., 2017). The Mycogen Corporation, a private peanut breeding program, has also developed a High-O/L ratio cultivar, ‘Flavor Runner 458’ (ASA et al., 2009) by chemical mutagenesis within the ‘Florunner’ (Norden et al., 1969) cultivar.
There are many enzymes regulated by gene(s) involved in fatty acid biosynthesis of linoleic from oleic (Gurr and James, 1975). Linoleic fatty acid (cis, cis -9, 12-18:2 or C18:2) is an essential fatty acid in humans and animals that must be supplied in the diet from plant sources like peanut. The major site of fatty acid biosynthesis in plants occurs in the chloroplast. This process needs light for photosynthesis to generate adenine triphosphate (ATP), which is supplied in the leaves and translocated to the developing peanut seed as stored energy reserves needed during germination. Because there are different sources of high-oleic peanut genotypes, the objective of this study was to use allelism tests between High-O/L x High-O/L and Very High-O/L x Very High-O/L cross combinations for determining possible differences among these high-oleic peanut genotypes.
Materials and Methods
During 2012-15, fatty acid methyl esters (FAME) were determined by gas chromatography (GC) as a percentage of the total oil composition. Ten sound mature kernels (SMK) of seven normal-oleic and seven high-oleic peanut cultivars were sent to the W. M. Keck Metabolomics Research Laboratory at Iowa State University. The oleic to linoleic (O/L) fatty acid ratio was used to separate genotypic differences. Each year the same 14 peanut cultivars were grown under maximum-input production practices with irrigation at the University of Georgia (UGA), Coastal Plain Experiment Station (CPES), Gibbs Farm near Tifton, GA.
A long growing season annually occurs at this location from the first of April through October. All genotypes were individually dug near optimum maturity based upon the hull-scrape method from adjacent border plants (Williams and Drexler, 1981). After harvest, peanut pods from each plot were dried with forced warm air to approximately 6% moisture. Pods were pre-sized and shelled on Federal-State Inspection Service equipment. Samples were then screened, and only sound mature seed were used to lessen any maturity effects for fatty acid determinations.
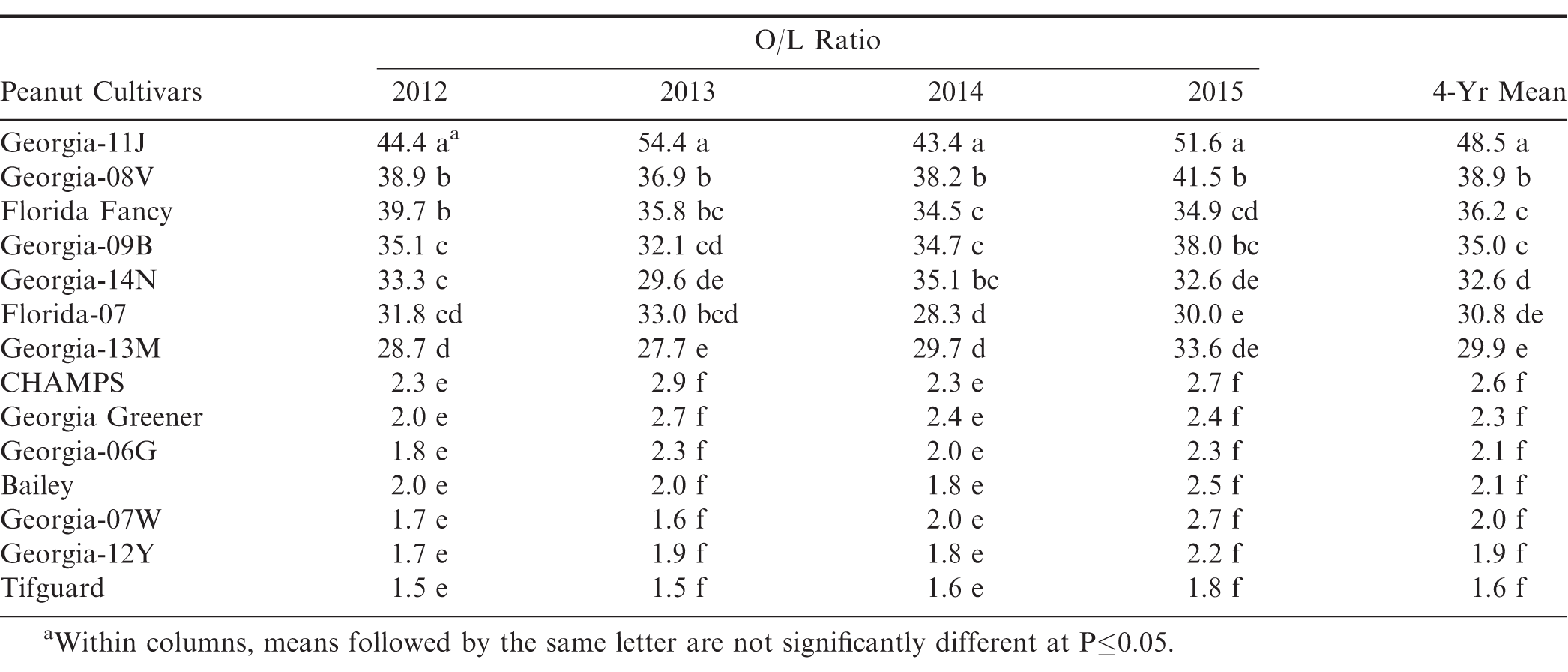
A randomized complete block field design was used for cultivar evaluations with five replications. Data from O/L ratios were subjected to ANOVA analysis of variance. Waller-Duncan Bayesian T-test (k-ratio = 100) was used for mean separation at P≤0.05 in SAS (SAS, Cary, NC) (Table 1).
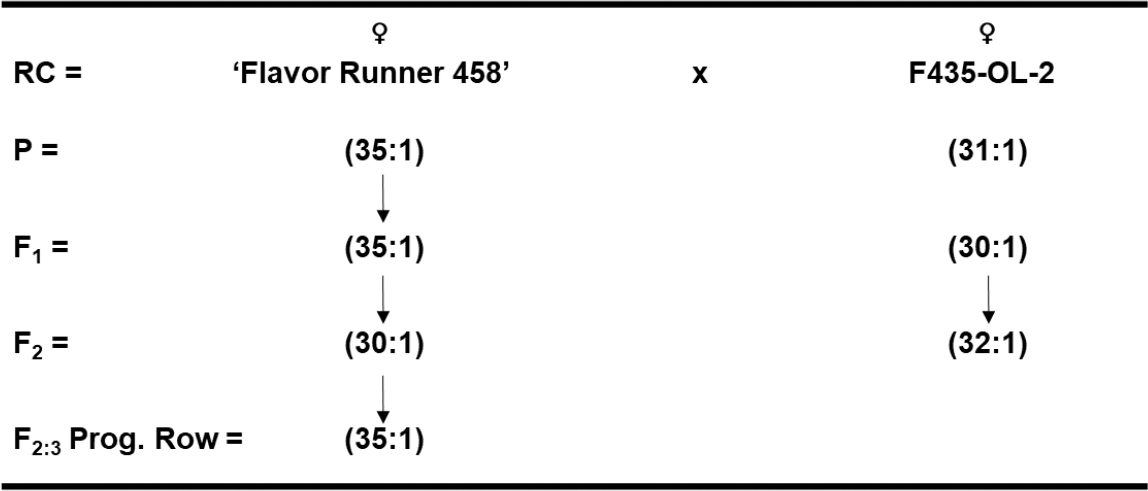
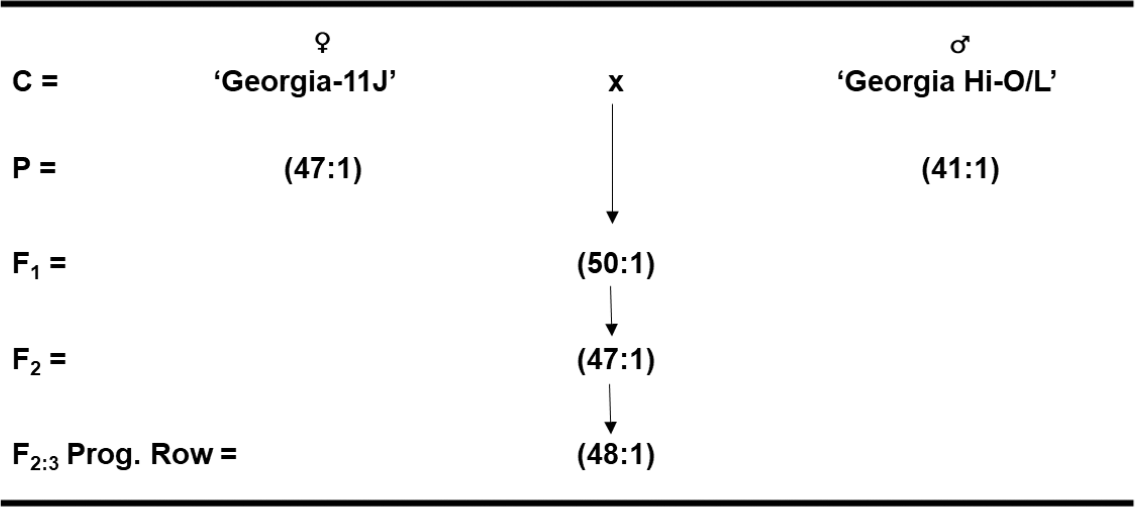
Four cross combinations were made in the greenhouse between two High-O/L parental lines, Flavor Runner 458 x F435-OL-2, and between two Very High-O/L cultivars ‘Georgia-11J’ (Branch, 2012) x Georgia Hi-O/L to test for possible allelic differences. Seed of each cross combination were space-planted 122-cm apart for F1 and 30.5-cm apart for F2 and F3 generations, respectively. Recommended cultural practices with irrigation were used throughout each growing season at this same UGA/CPES Tifton, GA location. These field nursery plots were in a three-year rotation following corn (Zea mays L.) and cotton (Gossypium hirsutum L.). Each year, plots consisted of two rows with variable length depending upon number of seed by 1.8 m wide beds, and were planted on a Tifton loamy sand soil type (fine-loamy, siliceous, thermic, Plinthic Kandidult). Individual plants were harvested near optimum maturity based upon days after planting and above-ground appearance. After harvest, peanut pods were dried with forced warm-air to approximately 6% moisture content before weighing and shelling. Ten SMK per plant were sent to W. M. Keck Metabolomics Research Lab for oleic and linoleic fatty acid methyl ester determination by GC from parents, F1, F2, and F3 cross populations.
Results and Discussion
Average oleic to linoleic (O/L) fatty acid ratio of fourteen peanut cultivars showed that the Georgia-11J cultivar had a Very High-O/L ratio > 40:1 significantly higher than the other High-O/L cultivars (20 to 40:1), which were all significantly higher than the normal-oleic cultivars (1 to 2:1) (Table 1). The Very High-O/L ratio of Georgia-11J was consistent across all four-years (2012-15) as was the Very High-O/L ratio of Georgia Hi-O/L consistent across three-years (1997-99) and two production practices (Branch et al., 2003).
Allelism test crosses between High-O/L x High-O/L peanut genotypes (Flavor Runner 458 x F435-OL-2) resulted in F1 hybrid seed with an O/L ratio of 35:1 and 30:1 for the reciprocal crosses, respectively (Fig. 1). The F2 population likewise had an averaged O/L ratio of 30:1 vs 32:1 for the reciprocal cross combination. So, there appears to be no reciprocal difference among these individuals, and thus a lack of maternal and cytoplasmic effect for O/L fatty acid ratios. The F2-derived progeny rows in the F3 generation averaged a 35:1 O/L ratio as well.
Similar to the parents, allelism test crosses between Very High-O/L x Very High-O/L peanut cultivars (Georgia-11J x Georgia Hi-O/L) resulted in F1 hybrid seed with an O/L ratio of 50:1 (Fig. 2). Likewise, the F2 populations had an average O/L ratio of 47:1, and the F2-derived progeny rows in the F3 generation averaged 48:1. These results suggest that there are very little allelic differences among similar parental lines for High-O/L x High-O/L cross combinations as well as Very High-O/L x Very High-O/L crosses. However, individual plant selections have been made within the latter Very High-O/L x Very High-O/L cross with even higher O/L ratios to see if these progeny rows breed true-to-type.
Poehlman (1959) defined multiple alleles as a series of alleles, or alternative forms, of a gene. He further stated that multiple alleles arise by repeated mutations of a gene, each mutant giving different effects. Since high-oleic peanut genotypes originated by mutations, then it only seems reasonable to assume that these different mutation events could have created different multiple alleles. The significant differences found among these very high-oleic and high-oleic peanut genotypes for percentage of oleic and linoleic fatty acid could be explained due to potentially different multiple alleles controlling the enzymes involved in peanut fatty acid biosynthesis.
Summary and Conclusions
The results from these allelism test crosses involving High-O/L x High-O/L and Very High-O/L x Very High-O/L parental lines suggest that there are at least two different types of high-oleic genotypes. Multiple-year analyses clearly show consistent and significant (P≤0.05) O/L ratio differences among peanut genotypes (Table 1). The reason for these differences between High-O/L and Very High-O/L could be attributed to possibly such multiple alleles or modifier genes coming from different genotypic sources controlling the high-oleic trait in the cultivated peanut. Plant selections will continue to be made within these Very High-O/L x Very High-O/L cross combinations for development of even higher potential pure-line Very High-O/L genotypes in the future.
Literature Cited
ASA, CSSA, and SSSA. 2009. Breeding for high oleic acid oils. CSA News 54(7): 4– 8.
Branch, W. D. 1991. Registration of ‘Georgia Runner’ peanut. Crop Sci. 31: 485.
Branch, W. D. 2000. Registration of ‘Georgia Hi-O/L’ peanut. Crop Sci. 40: 1823– 1824.
Branch, W. D. 2012. Registration of ‘Georgia-11J’ peanut. J. Plant Reg. 6: 281– 283.
Branch, W. D., Nakayama, T. and Chinnan. M. S. 1990. Fatty acid variation among U.S. runner-type peanut cultivars. J. Amer. Oil Chem. Soc. 67: 591– 593.
Branch, W. D., Brenneman, T. B. and Culbreath. A. K. 2003. Tomato spotted wilt virus resistance among high and normal O/L ratio peanut cultivars with and without irrigation. Crop Protection. 22: 141– 145.
Gurr, M. I. and James. A. T. 1975. Lipid Biochemistry: An Introduction (2nd Ed.)A Halstead Press Book, Chapman and Hall, London, and John Wiley & Sons, Inc., New York.
Isleib, T. G., Young, C. T. and Knauft. D. A. 1996. Fatty acid genotypes of five virginia-type peanut cultivars. Crop Sci. 36: 556– 558.
Knauft, D. A., Moore K. M., and Gorbet. D. W. 1993. Further studies on the inheritance of fatty acid composition in peanut. Peanut Sci. 20: 74– 76.
Moore, K. M. and Knauft. D. A. 1989. The inheritance of high-oleic acid in peanut. J. Hered. 80: 252– 253.
Mozingo, R. W., O’Keefe, S. F. Sanders, T. H. and Hendrix. K. W. 2004. Improving shelf-life of roasted and salted inshell peanuts using high oleic fatty acid chemistry. Peanut Sci. 31: 40– 45.
Nadaf, H. L., Biradar, K. Murthy, G. S. S. Krishnaraj, P. U. Bhat, R. S. Pasha, M. A. and Yerimani. A. S. 2017. Novel mutations in oleoyl-PC desaturase (ahFAD2B) identied from new high oleic mutants induced by gamma rays in peanut. Crop Sci. 57: 2538– 2546.
Norden, A. J, Lipscomb, R. W. and Carver. W. A. 1969. Registration of ‘Florunner’ peanuts (Reg. No. 2). Crop Sci. 9: 850.
Norden, A. J., Gorbet, D. W. Knauft, D. A. and Young. C. T. 1987. Variability in oil quality among peanut genotypes in the Florida breeding program. Peanut Sci. 14: 7– 11.
Poehlman, J. M. 1959. Breeding Field Crops. Holt, Rinehart, and Winston, Inc., New York, NY.
Williams, J. E. and Drexler. J. S. 1981. A non-destructive method for determining peanut pod maturity. Peanut Sci. 8: 134– 141.
Notes
- Professor, University of Georgia, Department of Crop & Soil Science, Coastal Plain Experiment Station, 2360 Rainwater Road, Tifton, GA 31793-5766 [^]
- Scientists, W. M. Keck Metabolomics Research Laboratory, Office of Biotechnology, Iowa State University, Ames, IA 50011-1010 [^] *Corresponding author Email: wdbranch@uga.edu
Author Affiliations