Introduction
Peanut (Arachis hypogaea L.) is the third major oilseed crop in the world next to soybean and cotton. However, drought which is the single most devastating environmental stress, not only decreases crop productivity (Lambers et al., 2008) but also predisposes peanut to Aspergillus sp. invasion and aflatoxin contamination making it unfit for consumption (Holbrook et al., 2000).
Soil is a reservoir for A. flavus, A. parasiticus and other species from section Flavi (Horn et al., 1995; Horn and Dorner 1998). In contrast to aerial crops, peanuts with their subterranean growth habit are exposed directly to soil populations of these fungi. Soils contains various types of microorganisms in the seed microenvironment (geocarposphere); however, these microorganisms do not invade the seed and produce aflatoxins until seed moisture content is lowered and the seed experiences long term drought stress, lose their phytoalexin producing ability and become more susceptible to fungal entry (Azaizeh et al., 1989). The relationship between soil population density and the incidence of seed colonization by Aspergillus sp. were studied by Horn (2006), who concluded that species of section Flavi more readily colonized peanut seed compared to other sections when the seed is wounded naturally or by insects.
Phytoalexins such as stilbenes are known as 'stress metabolites' (Langcake, 1981). Plants tend to respond to pathogen attack by up-regulation of certain metabolites for protection against the pathogen. This self-defense potential includes the synthesis and production of antimicrobial (Chalal et al., 2014) secondary metabolite phytoalexins, known as stilbenes that are composed of two phenol components linked by a C2 bridge (Jyske et al., 2016) and synthesized via the phenylalanine pathway (Hasan et al., 2013a). Comparatively, as per the reports, resveratrol shows moderate antimicrobial activity, and is the precursor of more active derivatives such as piceid, pterostilbene and ℇ-viniferin (Chong et al., 2009a; Jeandet et al., 2010; Adrian et al., 2013).
The presence of the stilbene resveratrol in peanut was first detected in the hypocotyls infected by Helminthosporium carbonum and implicated as a phytoalexin (Ingham, 1976). Research has also shown that aflatoxin production in kernels is mitigated when plants maintain high relative water content, allowing phytoalexin production. However, under prolonged drought conditions, phytoalexin production is curtailed in the seed, and low soil moisture and high temperature conditions favor Aspergillus invasion and growth (Dorner et al., 1989). Thus, drought is known to limit phytoalexin production, promote fungal invasion, alter plant and fungal metabolism and predispose peanut to aflatoxin contamination (Blankenship et al., 1984; Cole et al., 1989).
Since Aspergillus is known to invade peanut mainly under prolonged drought conditions (20-30 days before harvest, Sanders et al., 1985), screening for tolerance to this invasion must be conducted under water stress using a rainout shelter or man-made concrete bunker which is difficult and expensive. In addition, optimum geocarpospheric temperatures between 26.7 and 43.3C, with a relative humidity of 62-99% and moisture content of 13-20% (Sumner and Lee, 2009) has to be maintained to achieve Aspergillus growth and aflatoxin production. Hence, breeding for drought and/or Aspergillus tolerance to avoid aflatoxin contamination in peanuts is cumbersome and expensive because detection and quantification of aflatoxins is costly (Jeyaramraja et al., 2018) and demands major effort and resources.
Recently, deep sequencing analyses of the major peanut fungal pathogen Aspergillus flavus in response to resveratrol confirmed that these stilbenoids can affect the expression of A. flavus genes leading to abnormal mycelia development resulting in fungal inhibition (Wang et al., 2015). The phytoalexin content of peanuts can be increased by germination or elicited by microbial infection. Postharvest induction events such as soaking and drying, wounding, UV light exposure (Hasan et al., 2013b) or attack by insects (Sobolev et al., 2007) can also help in increasing the production of phytoalexins. Arora and Strange (1991) showed that when the seed are sliced and incubated, the induction of resveratrol and other stilbenes are higher compared to control intact seeds. The resveratrol content of peanut sprouts was also shown to increase when grown under 95% relative humidity at 25C in dark for 9 days (Wang et al., 2005) compared to un-germinated seed.
In view of these reports, this study attempted to determine the relationship between the ability of peanut seed to produce stilbenes and subsequent Aspergillus invasion to evaluate the potential for stilbenes quantification in large scale evaluations of breeding population under laboratory conditions in lieu of field studies for identifying Aspergillus tolerant genotypes to help accelerate the breeding program.
Material and Methods
Materials
Twenty peanut (Arachis hypogaea L.) genotypes with varying drought tolerance levels were obtained from Acharya N. G. Ranga Agricultural University, Andhra Pradesh, India, and used in the study. The identities of these genotypes are:DRT-40, DRT-43, K1504, K1622, Kadiri-7, K1620, K1814, Narayani, K1632, Tirupati-3, K1320, K1470, K1559, K1563, K1569, K1570, K1604, K1809, K1812, and Anantha. Preliminary studies were conducted using a commercial cultivar, Georgia Green (GG) to optimize stilbenes induction protocol and Aspergillus inoculation studies. The entire experiment was conducted at the Florida A&M University, Plant Biotechnology Lab, Center for Viticulture and Small Fruit Research, Tallahassee, Florida 32317.
Stilbene Induction
The peanut seeds (cv. Georgia Green) were soaked over-night in water to break seed dormancy. The following day, seeds were sliced (1 to 2 mm) using a sharp blade and transferred to Petri dishes lined with moistened tissue paper and kept in a desiccator in the dark at 90-95% relative humidity and 25C. The sliced peanut cotyledons were sampled in three replications at daily intervals for ten days, stilbenes were then extracted and quantified using HPLC to identify the incubation period that results in maximum stilbenes expression. The incubation period that resulted in highest stilbenes expression was used for evaluating peanut genotypes to determine differences in their ability to produce stilbenes and avoid Aspergillus spp. colonization as described below.
Peanut Germplasm Evaluation
The twenty peanut genotypes described under Materials were used for determining differences in their stilbenes expression ability and Aspergillus colonization levels. The seed were soaked overnight in water, sliced (1 to 2 mm), kept in a desiccator in dark at 90-95% relative humidity and 25C temperature and incubated for four days. The sliced peanut cotyledons were sampled in three replications after four days of incubation, and stilbenes were extracted and quantified using HPLC to quantify stilbenes expression and determine genetic variation in their stilbene expression level.
Stilbenes Quantification
The peanut cotyledon slices were ground to a powder using mortar and pestle under liquid nitrogen. A portion (0.5g) of the ground powder was extracted with ethyl acetate:methanol (50:50, v:v), centrifuged and filtered through a 0.22μm nylon membrane filter and subjected to High-Performance Liquid Chromatography (HPLC). HPLC analysis of stilbenes was carried out using a Waters HPLC system equipped with a 2487 dual UV detector and gradient pump (Waters 1525). The HPLC pumps, auto sampler, and detectors were controlled via Waters Empower software (Empower 3 service pack 2, Waters). The analytical column, Luna RP C18 (4.6 × 250 mm; particle size 5 μm), and its corresponding guard column were used (Phenomenex) to separate the stilbenes. The column temperature was maintained at 25C. The column was eluted using a solvent gradient of acetonitrile (Solvent A) and water (Solvent B) as follows: 0-18 min, 90% solvent B (water), 10% solvent A (acetonitrile); 18-23 min, 85% A, 15% B; 23-30 min, 85% A, 15% B and 30-35 min, 10% A, 90% B. Flow rate was 0.4ml/min. An aliquot of the filtered sample solution (2μL) was injected into HPLC and UV absorbance of eluting material was recorded at wavelengths of 285 and 305 nm in two separate channels to determine its stilbenes content and composition. Quantification of stilbenes was performed using a calibration curve generated from known quantities of stilbene standards, t-piceid, t-resveratrol, ℇ-viniferin and t-pterostilbene which were purchased from Sigma Chemical Co. (St. Luis, MO). Metabolite extraction and HPLC analyses were repeated at least three times to address biological as well as analytical variations. The concentration values, including standard error for each stilbene found in each genotype were analyzed using Microsoft Excel 2010.
Aspergillus flavus culture
Aspergillus flavus was purchased from the American Type Culture Collection (ATCC) and multiplied on Czapek Agar media prepared according to the published procedure (Czapek, 1902-1903). The spores were collected and quantified using Tryphan blue dye and cell counter (Bio-Rad). The concentration of viable cells was adjusted to 2.16 × 105 cells mL-1 and used in seed colonization studies described below.
Aspergillus flavus invasion studies
The resveratrol induction period (four-days) that was found (Materials and Methods: results of Stilbenes induction study above) to result in maximum stilbenes induction in peanut seed slices was used for fungal inoculation as follows. Stilbenes-induced and no-stilbenes induced (control) peanut seed slices were dipped in the diluted fungal spore suspension (2.16 × 105 cells mL-1) for five seconds individually, the excess liquid was shaken off, then slices were placed on Petri dishes. The inoculated seed slices were incubated at 32C and the fungal colonization level was visually monitored. Three controls were used to validate the observations: Control 1= Slices from no-stilbenes induced seed, Control 2= Slices from freeze-thawed seed, Control 3= Slices disinfected by dipping in 5% bleach.
Results and Discussion
Optimization of stilbenes induction protocol
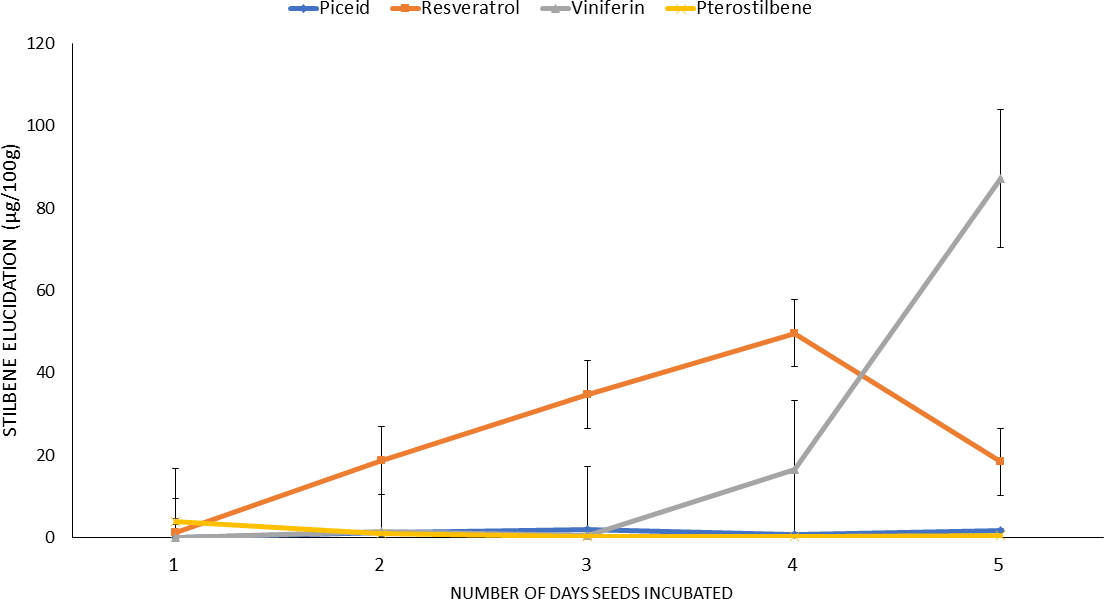
Initially, the stilbenes induction protocol was optimized using a commercial cultivar, Georgia Green and the optimized protocol was used in the rest of the study. The mature seed were soaked, sliced and initially incubated for up to ten days. However, the data showed that incubation beyond five days resulted in stilbenes loss and tissue decay; therefore, incubation was stopped after five days. The stilbenes analysis of the incubated seed extracts showed that peanut seed slices expressed mainly four stilbenes viz. t-piceid, t-resveratrol, ℇ-viniferin, and t-pterostilbene and their proportions differed significantly. Monitoring stilbenes content of the seed slices during the five-day incubation period showed presence of only trace levels of piceid in the seed which remained similar during rest of the incubation periods. In contrast, the resveratrol content of seed slices increased from 1.22 μg/100g to 49.64 μg/100g between day one and day four (Fig. 1) of incubation. However, on day five of incubation the resveratrol content decreased to 18.30 μg/100g. The ℇ-viniferin content of the seed slices remained negligible until day three and then gradually increased and reached its highest level(87.179 μg/100g) by day five of incubation (Fig. 1). Interestingly, the t-pterostilbene content of the seed decreased by day two and remained unchanged during rest of the incubation period. The control seeds viz. non-soaked, freeze-thawed and bleach treated seeds, failed to produce any stilbenes indicating that these seed slices were not elicited to produce stilbenes or incapable of producing any stilbenes, respectively. The above experiment was repeated multiple times (>6) to validate the observations and the results always showed maximum stilbenes induction by fourth day of incubation; hence, this period of incubation was used in the rest of the study. This result is consistent with the observation of Wang et al. (2005) who reported enhanced stilbenes expression in germinated seeds.
Genetic variation in stilbene expression among peanut genotypes
The 20 peanut genotypes with diverse drought tolerance characteristics obtained from ANGR Agricultural University were subjected to the optimized stilbenes induction protocol and the seed slices were harvested after four-days of incubation, then stilbenes were extracted and quantified using HPLC. The results showed that all 20 genotypes expressed four stilbenes viz. t-piceid, t-resveratrol, ℇ-viniferin and t-pterostilbene but their content greatly varied among the genotypes.
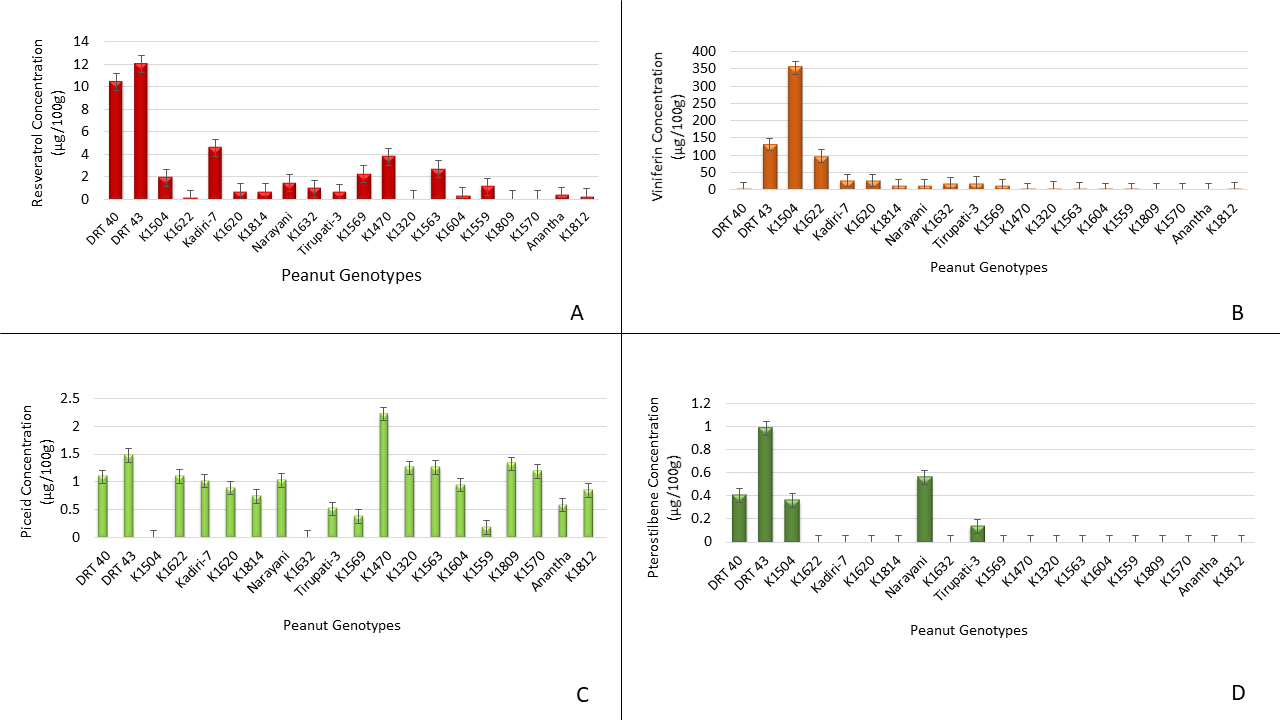
Comparative analysis of individual stilbenes of peanut genotypes showed that the genotype DRT 43 expressed highest amount of resveratrol (12.04 μg/100g tissue) followed by DRT 40 (10.5 μg/100 g tissue) while the genotypes K1320, K1809 and K1570 showed no resveratrol production (Fig. 2A). The other genotypes produced relatively lower amounts of resveratrol which were significantly lower than the resveratrol produced by genotypes DRT43 and DRT40. It is interesting to note that the genotypes DRT 43 and DRT 40 not only produce highest amounts of stilbenes but also are drought tolerant.
In addition to resveratrol, ℇ-viniferin expression also varied significantly among the genotypes (Fig. 2B). Among the genotypes studied, genotype K1504 expressed the highest amount of ℇ-viniferin (353.33 μg/100g) followed by DRT 43 (131.55 μg/100g) and K1622 (97.53 μg/100g) while the genotypes K1470, K1320, K1563, K1604, K1559, K1809, K1570, Anantha and K1812 produced none to trace amount of ℇ-viniferin. Unlike resveratrol and ℇ-viniferin, most genotypes produced t-piceid except for K1504 and K1632 (Fig 2C). Among the genotypes, K1470 produced the greatest amount of t-piceid (2.22 μg/100g) followed by DRT 43 (1.47 μg/100g), while the rest of the genotypes produced varying levels of t-piceid.
Since the stilbenes, piceid, viniferin and pterostilbene are analogs of resveratrol and share similar structure (Cichewicz and Kouzi, 2002), an increase in one leads to the decrease in level of the other. Thus, differences in stilbenes content of genotypes DRT40, DRT43, K1504 and Kadiri-7 is inversely proportional to their stilbene composition. Interestingly, only five peanut genotypes produced t-pterostilbene while others produced no t-pterostilbene. Among them DRT43 produced the highest amount of t-pterostilbene (0.98 μg/100g) followed by Narayani (0.56 μg /100g) (Fig. 2D). Overall the stilbenes analysis showed that genotypes DRT40, DRT43, Narayani and Tirupati-3 expressed all four stilbenes in varying amounts while the others failed to produce all four stilbenes which might impact their disease tolerance.
Based on the stilbenes content and composition, the 20 genotypes studied were grouped into High- and Low-stilbenes producing genotypes. According to the stilbenes content and composition, five high- and five low-stilbenes producing genotypes were selected and used in the Aspergillus inoculation study described above to determine the relationship between stilbenes content and composition and Aspergillus colonization of the seed. Therefore, DRT40, DRT43, K1504, K1622 and Kadiri-7 were considered as high-stilbenes producing genotypes while K1620, K1814, Narayani, K1632 and Tirupati-3 were taken as low-stilbenes producing genotypes.
Relationship between stilbenes content/composition and Aspergillus flavus colonization of peanut genotypes
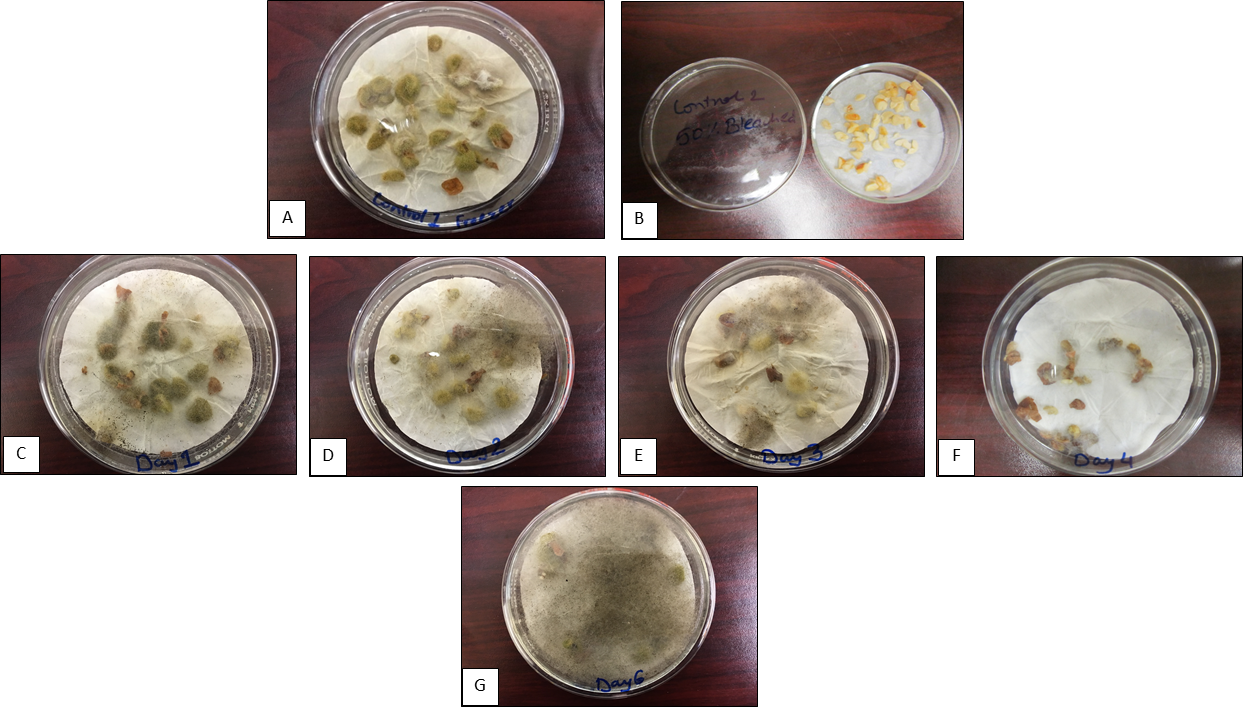
The Aspergillus colonization study was optimized initially using a commercial cultivar Georgia Green. The water soaked and sliced seeds were inoculated after each day of incubation for up to five days with an Aspergillus flavus spore suspension (2.16 × 105 cells mL-1) and were incubated for additional seven days while fungal colonization of the slices were monitored. The results showed (Fig. 3) that seed slices inoculated one- and two-days after incubation were heavily colonized by the fungi while the ones inoculated beyond two days of incubation showed decreasing colonization levels reaching to minimal colonization by day four of incubation. Additional seed incubation periods beyond four days and inoculation with fungus enhanced fungal colonization and resulted in seed decay by day six (Fig. 3). It is interesting to note that stilbenes expression in the seed gradually increased from day one of incubation and reached a maximum by day four (resveratrol) and day five (ℇ-viniferin) of incubation (Fig. 1) indicating that with increasing levels of stilbenes expression fungal colonization decreased (Fig. 3). However, as the stilbenes expression decreased, the fungal colonization level increased after five days of incubation. These data (Fig. 1 and Fig. 3) clearly demonstrated a negative correlation between stilbenes content and Aspergillus colonization levels of the seed.
Progressive changes in Aspergillus colonization level of the cotyledon slices at different incubation periods. Control A= Freeze-thawed seed, Control B= Seed treated with 5% bleach, C= Day 1, D= Day 2, E= Day 3, F= Days 4, G= Day 6 of incubation. Note progressive changes in Aspergillus fungal colonization level between the incubation periods C to G.
The observation that stilbenes expression must precede fungal inoculation to prevent colonization was further confirmed using multiple seed lots and replications using unincubated seed as controls. The water soaked peanut seed was sliced, incubated for four days to induce maximum stilbenes expression and then inoculated with Aspergillus to assess the effect of stilbenes on fungal colonization level. The unincubated seed (no stilbenes expressed), freeze-thawed seed and bleach disinfected seed were used as controls to demonstrate that stilbenes expression is a pre-requisite for preventing fungal colonization. The results showed that the control seed was colonized by Aspergillus while the incubated seed showed no Aspergillus colonization (Fig. 4). The seed from both the treatments was analyzed for stilbenes and found that four-day incubated seed expressed stilbenes while the control seeds were devoid of any stilbenes. These data clearly demonstrated that the four-day incubated seed slices with stilbenes were able to overcome fungal colonization while the unincubated/freeze-thawed/bleach disinfected control seeds devoid of stilbenes and succumbed to fungal colonization. These data further confirm that the seeds with stilbenes were able to avoid fungal colonization while the ones without stilbenes were colonized by Aspergillus.
Validation of Aspergillus invasion avoidance by four-day incubated seed due to their high stilbenes content. Peanut seeds (cv. Georgia Green) were soaked in water, sliced (1 to 2 mm) and incubated in a desiccator at 25C for four days and then inoculated with Aspergillus spore suspension (2.16 × 105 cells mL-1), A= Control-unincubated seed, B, C and D= Peanut seed slices were incubated for four days, inoculated with Aspergillus spore suspension and incubated for additional 7 days to view fungal colonization. Note little to no Aspergillus colonization of incubated seed while unincubated seed were heavily colonized by the fungus.
Differential response of peanut genotypes to Aspergillus colonization
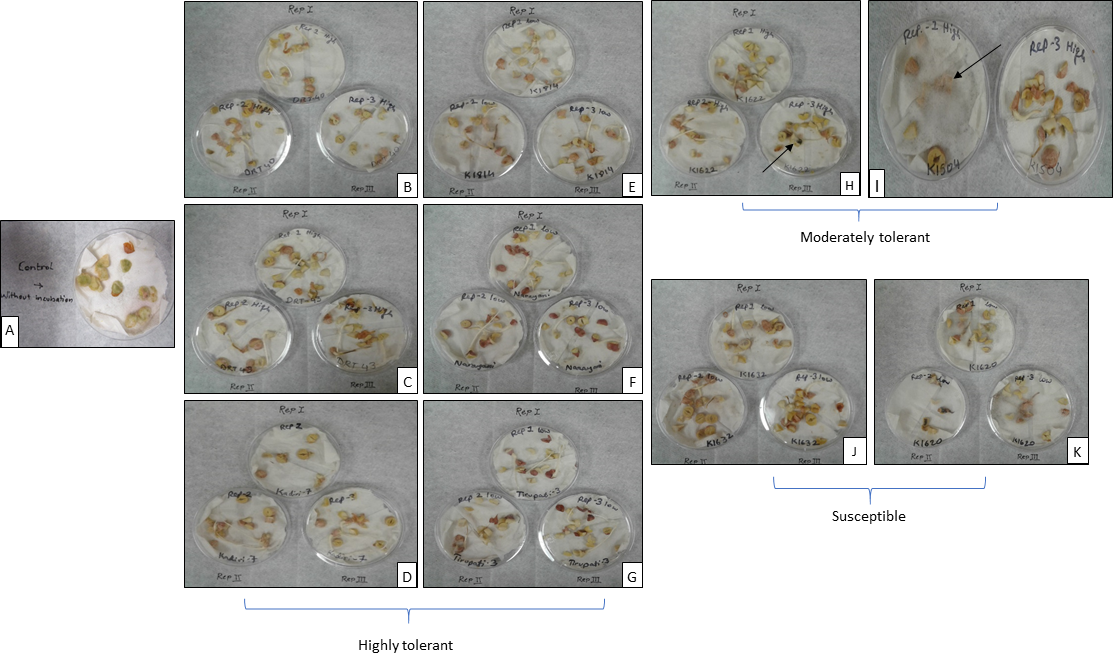
To determine the potential of peanut genotypes to evade Aspergillus colonization, peanut genotypes with varying stilbenes producing ability were tested for susceptibility to Aspergillus colonization. The peanut genotypes with varying stilbenes producing ability were selected based on their ability to express stilbenes (Fig. 2) and used to measure their capability to avoid Aspergillus colonization. The results showed (Fig. 5) that peanut genotypes that generally expressed higher amounts of stilbenes showed no fungal colonization while the genotypes below a threshold amount of stilbenes were colonized with Aspergillus. Thus, the genotypes DRT40, DRT43 and Kadiri-7, which produced high amounts of stilbenes, were able to mitigate the fungus, while moderate-stilbenes producing genotypes were also resistant to fungal colonization (Fig. 5). However, the low stilbenes producing genotypes such as K1620 and K1632 showed heavy fungal colonization. In addition, the stilbenes composition also seems to play a role in fungal tolerance level. For example, genotype DRT43 contained high amounts of all the four stilbenes, while DRT40 contained high amounts of resveratrol, piceid and pterostilbene, and only trace amounts of viniferin, but was still able to resist fungal colonization (Fig. 5). Interestingly, the genotypes K1814 and Tirupathi 3 contained lower amounts of resveratrol, viniferin and pterostilbene but a higher amount of piceid, and was able to withstand fungal colonization. Likewise, Narayani contained lower amounts of resveratrol and viniferin but high levels of piceid and pterostilbene and avoided fungal invasion. While, the genotype K1504 contained highest levels of ℇ-viniferin among the genotypes tested, it produced very low levels of resveratrol and pterostilbene, and no piceid, and was susceptible to Aspergillus colonization (Fig. 5). These results suggest that although total stilbenes content is an important component to avoid fungal invasion, stilbenes composition is also important. This is expected since the stilbenes are analogs of resveratrol and known to possess different toxicity levels (Chong et al., 2009b). Overall, the outcome of this study revealed that peanut seed that produce above threshold amounts of stilbenes avoid fungal invasion and potential aflatoxin contamination as evidenced by avoidance of Aspergillus colonization by high stilbenes producing varieties such as DRT40, DRT43 and Kadiri-7.
Interrelationship between stilbenes content and composition, and Aspergillus colonization level among peanut genotypes with varying stilbenes production capability. Seed from different peanut genotypes with varying stilbenes producing ability were soaked in water, sliced, incubated for four-days at 25C, inoculated with Aspergillus and further incubated for seven days. A=Control: without incubation, B= DRT40, C=DRT43, D= Kadiri7, E=K1814, F=Narayani, G= Tirupati 3, H= K1622, I= K1504, J=K1632, K= K1620. Note none to minimal Aspergillus colonization in genotypes A to G and colonization in genotypes H to K.
Conclusion
This study attempted to prove that peanut genotypes capable of producing above threshold amounts of stilbenes would avoid Aspergillus invasion. Results show that peanut genotypes that produce suitable amounts and types of stilbenes were able to overcome Aspergillus invasion and potential aflatoxin contamination. In addition, the data also showed that besides stilbenes level, the composition is also important since certain stilbenes combinations were found to be more effective in preventing fungal colonization than others. In addition, the genotypes that express mostly resveratrol and piceid appear to be more effective in avoiding fungal invasion than those that produce other stilbenes. The in vitro stilbenes induction and fungal colonization techniques used in this study are simple, less expensive compared to existing techniques and may be useful for large scale screening of breeding population to identify Aspergillus-tolerant genotypes and accelerate the breeding program.
Acknowledgements
This research was funded by a grant from USDA-NIFA, Evans Allen and Capacity Building Grant programs. The authors report no conflict of interest.
Literature Cited
Adrian, M., M De Rosso, L Bavaresco, B Poinssotand M.-C Héloir (2013) Resveratrol from vine to wine, Nova Science Publishers Inc.: New York, NY, USA : 3-19.
M Arora, and R Strange ((1991)). "Phytoalexin accumulation in groundnuts in response to wounding.". Plant Science 78 ((2)): 157- 163.
H., Azaizeh, R Pettit, O Smithand R Tabef ((1989)). "Reaction of peanut genotypes under drought stress to Aspergillus flavus and A. parasiticus.". Peanut Science 16 ((2)): 109- 113.
P. D., Blankenship, R. J Cole, T. H Sandersand R. A Hill ((1984)). "Effect of geocarposphere temperature on pre-harvest colonization of drought-stressed peanuts by Aspergillus flavus and subsequent aflatoxin contamination.". Mycopathologia 85 ((1-2)): 69- 74.
M., Chalal, A Klinguer, A Echairi, P Meunier, D Vervandier-Fasseurand M Adrian ((2014)). "Antimicrobial activity of resveratrol analogues.". Molecules 19 ((6)): 7679- 7688.
J., Chong, A Poutaraudand P Hugueney ((2009)). "Metabolism and roles of stilbenes in plants". Plant science 177 ((3)): 143- 155.
R. H Cichewicz, and S. A Kouzi ((2002)). "Resveratrol oligomers: structure, chemistry, and biological activity". Studies in natural products chemistry, Elsevier 26: 507- 579.
Cole, R., T Sanders, J Dornerand P Blankenship (1989) Environmental conditions required to induce preharvest aflatoxin contamination of groundnuts: summary of six years' research International Workshop on Aflatoxin Contamination of Groundnut, Patancheru, AP (India), 6-9 Oct 1987, ICRISAT.
Czapek, F (1902/1903). "Untersuchungen uber die stickstoff gewinnung und Eisessbildung der Pflanzen." Beitr. Chem Physiol. u. Pathol . 1:540-560; 3: 47- 66.
J. W., Dorner, R. J Cole, T. H Sandersand P. D Blankenship ((1989)). "Interrelationship of kernel water activity, soil temperature, maturity, and phytoalexin production in preharvest aflatoxin contamination of drought-stressed peanuts.". Mycopathologia 105 ((2)): 117- 128.
M. M., Hasan, M Cha, V. K Bajpaiand K.-H Baek ((2013)). "Production of a major stilbene phytoalexin, resveratrol in peanut (Arachis hypogaea) and peanut products: a mini review.". Reviews in Environmental Science and Bio/Technology 12 ((3)): 209- 221.
C., Holbrook, C Kvien, K Rucker, D Wilson, J Hookand M Matheron ((2000)). "Preharvest aflatoxin contamination in drought-tolerant and drought-intolerant peanut genotypes.". Peanut Science 27 ((2)): 45- 48.
Horn, B and J Dorner (1998) "Soil populations of Aspergillus species from section Flavi along a transect through peanut-growing regions of the United States." Mycologia : 767-776.
B., Horn, R Greeneand J Dorner ((1995)). "Effect of corn and peanut cultivation on soil populations of Aspergillus flavus and A. parasiticus in southwestern Georgia.". Applied and Environmental Microbiology 61 ((7)): 2472- 2475.
B. W Horn, ((2006)). "Relationship between soil densities of Aspergillus species and colonization of wounded peanut seeds.". Canadian journal of microbiology 52 ((10)): 951- 960.
J. L Ingham, ((1976)). "3, 5, 4"-Trihydroxystilbene as a phytoalexin from groundnuts (Arachis hypogaea).". Phytochemistry 15 ((11)): 1791- 1793.
P., Jeandet, B Delaunois, A Conreux, D Donnez, V Nuzzo, S Cordelier, C Clémentand E Courot ((2010)). "Biosynthesis, metabolism, molecular engineering, and biological functions of stilbene phytoalexins in plants.". Biofactors 36 ((5)): 331- 341.
P., Jeyaramraja, S. N Meenakshiand F Woldesenbet ((2018)). "Relationship between drought and preharvest aflatoxin contamination in groundnut (Arachis hypogaea L.).". World Mycotoxin Journal 11 ((2)): 187- 199.
Jyske, T., K Kuroda, J.-P Suuronen, A Pranovich, S. R Juan, D Aokiand K Fukushima (2016) "In planta localization of stilbenes within Picea abies phloem." Plant physiology: pp 00990.02016.
Lambers, H., F. S Chapinand T. L Pons (2008) Photosynthesis Plant physiological ecology, Springer: 11-99.
P Langcake, ((1981)). "Disease resistance of Vitis spp. and the production of the stress metabolites resveratrol, ℇ-viniferin, 𝜶-viniferin and pterostilbene.". Physiological Plant Pathology 18 ((2)): 213- 226.
T. H., Sanders, R. J Cole, P. D Blankenshipand R. A Hill ((1985)). "Relation of environmental stress duration to Aspergillus flavus invasion and aflatoxin production in preharvest peanuts.". Peanut Science 12 ((2)): 90- 93.
V. S., Sobolev, B. Z Guo, C. C Holbrookand R. E Lynch ((2007)). "Interrelationship of phytoalexin production and disease resistance in selected peanut genotypes.". Journal of agricultural and food chemistry 55 ((6)): 2195- 2200.
Sumner, P. E and R. D Lee (2009) "Reducing aflatoxin in corn during harvest and storage." https://athenaeum.libs.uga.edu/handle/10724/12104.
H., Wang, Y Lei, L Yan, K Cheng, X Dai, L Wan, W Guo, L Chengand B Liao ((2015)). "Deep sequencing analysis of transcriptomes in Aspergillus flavus in response to resveratrol.". BMC microbiology 15 ((1)): 182.
K.-H., Wang, Y.-H Lai, J.-C Chang, T.-F Ko, S.-L Shyuand R. Y.-Y Chiou ((2005)). "Germination of peanut kernels to enhance resveratrol biosynthesis and prepare sprouts as a functional vegetable.". Journal of agricultural and food chemistry 53 ((2)): 242- 246.
Notes
- Center for Viticulture and Small Fruit Research, Florida A&M University, 6361 Mahan Drive, Tallahassee, FL 32308. [^]
- Agricultural Research Station, Kadiri - 515 591, Acharya N G Ranga Agricultural University, Kadiri, Andhra Pradesh, India. [^] *Corresponding author email; mehboob.sheikh@famu.edu
Author Affiliations