Introduction
Peanut is an important cash crop valued for its oil, protein, and flavor content. In 2016, 1.7 million acres of peanut were planted in the U.S. (National Agricultural Statistics Service, 2016). U.S. peanut yield achieved a six-fold increase from 739 kg ha−1 in 1909 to 4695 kg ha−1 in 2012 (Holbrook et al., 2014). Although yield increase is the outcome of advances in multiple aspects of agricultural practices, cultivar improvement played a critical role in this yield gain (Holbrook, et al., 2014). Cultivar development also introduced many desirable characteristics for peanut farming and industry such as integrated multiple disease and pest resistances, improved drought tolerance, and improved seed oil composition, etc. (Isleib, et al., 2001; Chu, et al., 2011). Three main approaches have been applied in crop improvement: 1) artificial hybridization commonly known as crossing, 2) mutagenesis by chemicals or radiation, and 3) genetic transformation. The second and third methods are not widely used in peanut breeding programs due to either difficulty in trait selection or economic feasibility. Artificial hybridization has given rise to most of the released peanut cultivars in the U.S. Furthermore, artificial hybridization has been employed in the effort to identify genetic components conferring phenotypic traits of interest. Current release of peanut diploid genome sequences (Bertioli et al., 2016) and tetraploid transcriptome data (peanutbase.org) vastly improves the opportunity of single nucleotide polymorphism (SNP) discovery among cultivated peanut varieties. However, to establish gene-trait associations for molecular marker development, mapping populations segregating for traits of interest are needed. Prior to 2011, 45 genetic mapping populations were established world-wide resulting in the discovery of quantitative trait loci (QTLs) associated with disease resistance, abiotic stresses and various agronomic traits (Pandey et al., 2012).
As is typical of species in the legume family, peanut flowers self-pollinate. Each flower has one large standard, two lateral wings, and a keel, which encloses the staminal tube at the distal end of which extends eight anthers surrounding a club-shaped stigma (Smith, 1950). Anther dehiscence and self-pollination occur during floral expansion shortly after sunrise. During pollination, mature pollen first attaches to the serrated tip of the stigma and hydrates with exudate from the stigma. With the supply of nutrients such as amino acids, carbohydrates, and lipids from the stigma, pollen germinates and pollen tubes travel down the style enclosed by the hypanthium to reach the ovary located at the base of the style (Swanson, et al., 2004). Sperm and egg unite to form a zygote that divides to initiate the embryo, which becomes quiescent after only a few cell divisions. Meristem activity at the base of the ovary causes an elongating peg to form 5 to 14 days after fertilization and elongate geotropically. Peanut embryos located at the tip of the peg resume development and a pod forms after the peg penetrates into the soil.
In order to create artificial hybrids, flowers from the female plants must be emasculated prior to anther dehiscence. Mature pollen released from male plants should be applied to the stigma of female plants. Hybrid pegs produced from the cross need to be identified and tagged before harvest. Since peanut artificial hybridization is low yielding and time consuming, reported to cost 10 minutes per flower (Hammons, 1964), maximizing the success rate of artificial hybridization is desirable for peanut breeding programs. Previously, 70-90% of hand pollinations were reported to achieve fertilization (Norden and Rodriguez, 1971) and 26% to 89% of pollinations resulted in viable hybrids (Banks, 1976). In these earlier crossing studies, inheritance of dominant phenotypic variation such as leaf shape, stem color and texture, and growth habit were used to distinguish hybrids from self-pollination derived F1 progenies (Banks, 1976) and the success rate was reported without statistical replicates.
Success rates of artificial hybridization can be affected by multiple factors such as humidity, temperature, crossing schedule, peanut genotype, operators and integrity of emasculated flowers etc. High humidity had also been shown to enhance peanut flowering and peg formation (Lee et al., 1972). Very low success rate of field hybridization was reported in the dry season in India and low humidity was possibly responsible for the low success rate (Norden, 1980). High temperature (> 33 C) had a negative impact on pollen grain number, pollen germination, pollen tube elongation and peg formation during three sensitive phases of peanut flower development, i.e. 1) microsporogenesis, which is 3-6 days prior to anthesis; 2) anthesis; and 3) pollination and fertilization (within 6-8 hr after anthesis) (Vara Prasad et al., 2001; Vara Prasad et al., 2002). Conventionally, emasculation is performed in the evening (17:00 to 19:00 hr) and cross-pollination the next morning between 7:00 to 10:00 hr (Norden and Rodriguez, 1971). This time schedule was modified previously by reversing day and night schedule of female flowers in a growth chamber (Banks, 1976) or delaying the female bud development by extending daylight time using an artificial light source in the greenhouse (Hildebrand, 1974). These modified schedules allow emasculation and pollination to occur at the same time in the morning. Although this schedule change is beneficial for performing crosses during regular work hours, the cost of growth chambers and modification of greenhouse settings can be a limiting factor for breeders. Success of an alternative concurrent pollination schedule by pollinating female flowers right after emasculation in the evening was suggested (Roy Pittman, pers. comm.). This concurrent pollination schedule was tested in comparison with a conventional schedule in our environment. Crossing peanut flowers by hand is a highly delicate job requiring skilled operators. The performance of operators, therefore, affects the success rate of artificial hybridization. Success rate of hybridization could also be affected by the flower integrity. The emasculated flowers can be damaged and fail to produce viable pegs. One way to check overall damage to the female flowers upon emasculation is to monitor the blooms in the morning. The success rate of emasculated flowers that opened fully in the morning was compared to those that failed to open. Another aspect that affects the success of artificial hybridization involves the selection of appropriate parents and identification of F1 hybrids. Peanut breeding lines are commonly advanced in the field where outcrossing can occur at 0.09 to 8% (Culp et al., 1968; Stone et al., 1973; Knauft et al., 1992). It is therefore possible that genetic heterogeneity can be found in the parental seed stock. Before the implementation of molecular markers, the level of heterogeneity introduced into crosses from parental lines was difficult to assess. Determination of F1 hybridity was conventionally performed by visual selection of dominant phenotypic traits transferred from male parents. However, in cases of no apparent visual markers, seeds from self-pollination can be mixed with hybrids, resulting in increased cost and difficulty of subsequent generation advancement and selection. With the most recent development of genetic markers, homozygous parental lines for the traits of interest can be selected prior to crossing and F1 hybridity can be checked upon seed germination (Favero et al., 2006; Chu et al., 2011). The objective of this study was to determine the effect of the factors described above on success rate of artificial hybridization. The percentage of single vs double-seeded pods among hybrids was also examined using runner type peanuts for crossing.
Materials and Methods
In the first experiment, common male parents for all female parents were selected from F3 plants of C1976 [Tifguard ((Holbrook et al., 2008) × (Tifguard × (Tifguard × (Tifguard × Florida-07 (Gorbet et al., 2009)))] that were genotyped to be homozygous for both high oleic acid (O/L) and nematode resistance traits by molecular markers developed previously (Chu et al., 2011). Briefly, genomic DNA was extracted from young, folded peanut leaf tissue by a high-throughput method (Xin et al., 2003). Primer set GM565F: 5′-TTTCCTTTCAACCCTTCGTG-3′ and GM565R: 5′-AATGAGACCAGCCAAAATGC-3′ (Eurofins, Louisville, KY) was used to genotype for nematode resistance. The high O/L trait was genotyped by a high-resolution melting curve analysis using a Roche LightCycler (Roche Applied Sciences, Indianapolis, IN). AhFAD2 was amplified by primers (sense: 5′-TTTGACCCTTCACTCTTGTCTATTA-3′ and antisense: 5′-TCCCTGGTGGATTGTTCATGTA-3′) using LightCycler genotyping master mix (Roche Applied Sciences). The 441_442insA mutation in the ahFAD2B allele was detected by a sensor HybProbe (5′-CCAACACAGGTTCCCTCAGAC-3′) and an anchor probe (5′-CAACACAGGTTCCCTCAGAC-3′). Crosses included cultivars and breeding lines paired as described in Table 1 with four replicates of each pair.
Parental seeds were sown in 12 inch pots with a mix of 50% Promix (Premier Tech Horticulture, Quaker, PA) and 50% steam-sterilized sandy soil from the Coastal Plain Experiment Station in Tifton, GA. The six female genotypes were arranged on the bench top in a greenhouse as indicated in Fig. 1A. All replications of a genotype were arranged in a consecutive manner. Two HOBO humidity and temperature data loggers (Onset Computer Corporation, Bourne, MA) recording data every 30 min were positioned either close to the cooling pad (logger 1) or at the center of the greenhouse (logger 2). Three groups of operators consisting of two persons per group were assigned to this experiment. Group ID and schedule of pollination were tracked by colors of threads gently knotted on the calyx tube of female flowers. If an emasculated flower fully opened the next morning, a second thread of the same color was placed on the extended hypanthium. At the time of peg extension, dried flowers from the tip of pegs were collected and hypanthium length was measured. Two pollination schedules were tested, i.e., separate and concurrent schedules. For the separate schedule, the flower buds were emasculated between 17:00 to 19:00 hr the day before blooming and pollinated with pollen grains from fresh blooms on male plants the following day at 8:30 hr. For the concurrent schedule, flowers from the male plants were collected at 8:30 hr and saved at 4 to 8 C in a petri dish with a moistened paper towel. Right after emasculation between 17:30 to 19:00 hr, the flower buds were pollinated with the pollen grains from male flowers collected earlier on the same day. Half of the female flowers from each pot were pollinated with the separate schedule and the other half were pollinated with the concurrent schedule. To avoid confounding effects between genotype and operator, the three groups rotated around the bench every day so that each group worked on every genotype at the same frequency. Success rate of a cross combination was determined by the number of wired pegs divided by the total number of emasculated flowers.
To separate the effects of location and genotype on the success of crossing, a second experiment was performed with three breeding lines (Table 1) as female parents and each breeding line had six replicates. The common male parents were selected from F4 lines of C1805 (Tifguard x Florida07) homozygous for the high O/L trait. Arrangement of these female plants in the greenhouse is shown in Fig. 1B. Two sets of all genotypes were clustered either around data logger 1 or data logger 2. Each set consisted of three replicates of each genotype and they were distributed randomly within each cluster.
In a third experiment, all pods from three crosses (Table 1) were harvested to calculate the percentage of single-seeded pods in the hybrids versus self-pollination derived pods. Both hybrid and seeds from self-pollinations were genotyped with either high O/L or nematode resistance markers.
Emasculation and pollination procedures followed a previously published method (Norden and Rodriguez, 1971). A piece of cotton thread was tied on the hypanthium of the bud to mark the emasculated flower. To perform pollination, pollen grains from a male flower were squeezed onto a pair of flat-ended tweezers and transferred to the tip of the stigma on an emasculated female flower. Any blooms on a female plant that were not emasculated were removed in the morning to avoid self-pollination, and minimize the chance of misidentifying hybrid pegs. Emergence of a hybrid peg was identified by the presence of cotton thread on the attached withered flower 5-14 days after pollination. A wire (www.grainger.com) made of a pliable plastic thread with copper core was used to mark the position of the peg in a pot initially. As the peg extended, the wire was attached to the peg permanently by bending the wire and letting the peg go through the circle on the top of the wire. After crossing on a plant was completed, a handful of gypsum was applied to each pot to enhance peg strength and pod yield. Hybrid pods were harvested 60-70 days after peg formation. F1 hybrid seeds were genotyped with molecular markers described above.
Data analysis was performed by ANOVA using SAS 8.2 statistical software (SAS, Cary, NC). Tukey's test was performed to separate the means. Significant differences were detected at a P-value less than 0.05.
Results and Discussion
Parents of crosses were genotyped using molecular markers prior to crossing. In the first experiment, 19 potential male plants from F3 plants of C1976 [Tifguard × (Tifguard × (Tifguard × (Tifguard × Florida-07))] were genotyped and all of them were homozygous for both high O/L and nematode resistance markers. Tifguard is the donor of the nematode resistance trait and Florida-07 is the donor of the high oleic trait. Earlier generations of these plants had been selected for both traits by molecular markers; therefore, consistent inheritance of both traits was expected. In the second experiment, 2 out of 28 potential male plants from F4 lines of C1805 (Tifguard x Florida-07) were eliminated from the study since they lacked both molecular markers. In the third experiment, all parents demonstrated expected genotypes with our markers. Although the presence of off-type parents in a breeding program is rare, excluding off-type parents is critical to preserve the purity of hybrid lineages. Currently, limited markers are available to peanut breeders. Advances in peanut genome sequencing (Bertioli et al., 2016) and mapping efforts should produce more markers to assist breeding.
In the first experiment, a total of 119 putative hybrid pegs were tagged. Thirty-two pegs failed to produce viable seeds. Peg damage, pod rot and seed immaturity contributed to the loss. Upon pollination, it takes approximately 5-14 days for a hybrid peg to emerge and another 3-4 days before the peg can be permanently wired. It takes 10 to 14 days from wiring of the peg until the peg tip enlarges sufficiently to secure its position in the soil. Wired pegs can be damaged incidentally while emasculating or checking for newly formed pegs. A few rotten seeds were found and several pods yielded extremely shriveled seeds. One hundred ten putative hybrid seeds were treated to break dormancy and nine seeds failed to produce viable plants. All of the germinated plants were genotyped with both high O/L and nematode resistance markers. Eighty-nine F1 plants (81%) were unequivocally identified as hybrids. Twelve plants (10.9%) were identified as self-pollination derived. During harvest, two wires were noted as having two pods each. It was apparent that pegs from self-pollinated flowers grew into the holes of the wires marking hybrid pegs. Since we have molecular markers to distinguish self-pollination derived versus hybrid genotypes, they were harvested to maximize the chance of recovering hybrids. The distribution of pod losses and self-pollination derived seeds was random among treatment groups, thus data analysis of crossing success rate was based on wired peg numbers.
Pollination schedule
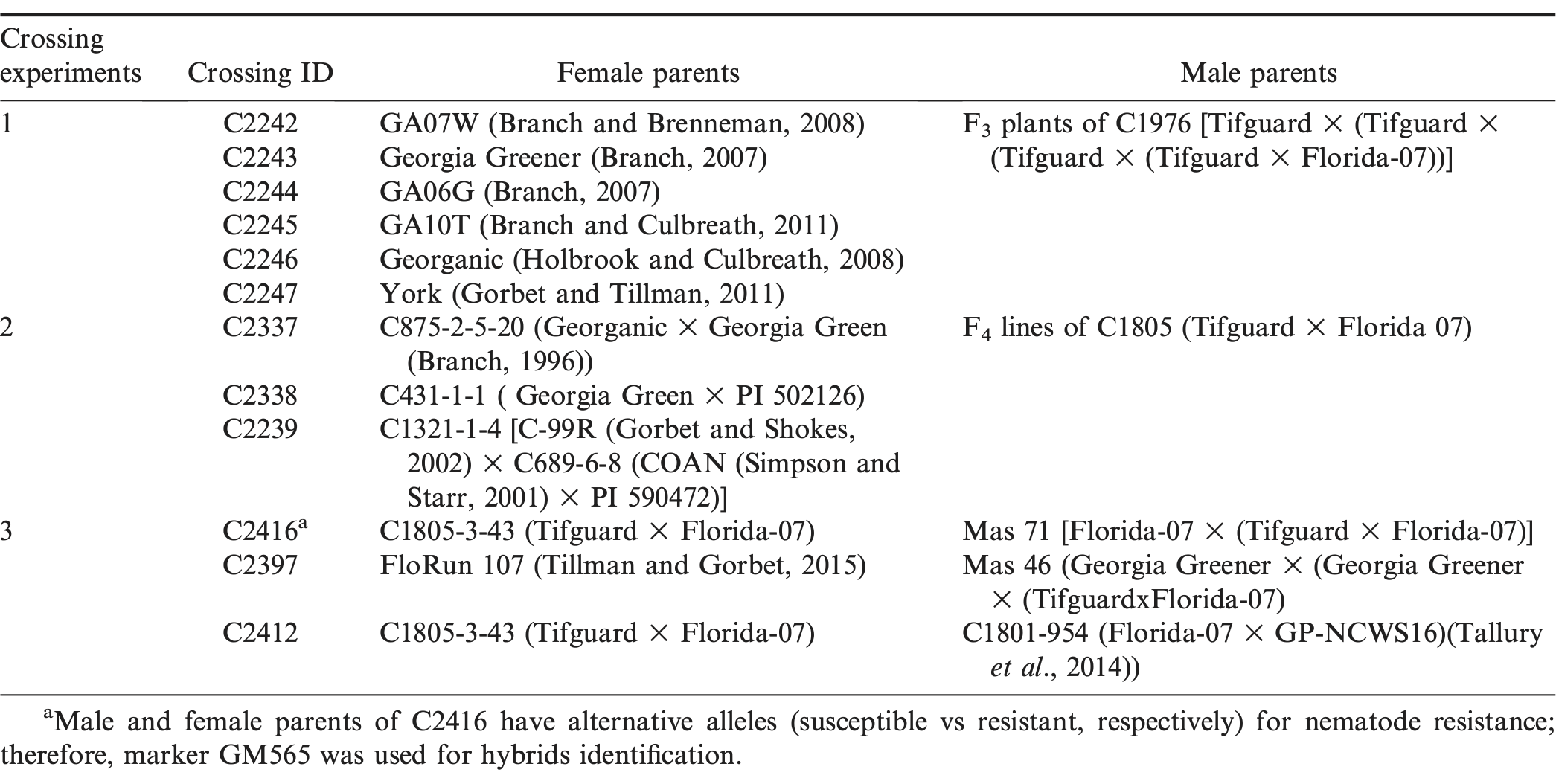
The “separate” pollination schedule produced significantly more pegs than that of the “concurrent” pollination schedule (Fig. 2A). Previously, pollen germination rate after storage at 8 C for one day was found to be 91% (Faucette and Emery 1974); therefore, it is unlikely that the lower success rate for the concurrent pollination schedule was due to the quality of pollen. Between 17:30 to 19:00 hr, the emasculation phase, the peanut flower bud was small and the stigma was short and thin which made it difficult to pollinate following the concurrent schedule. After pollination, the banner and wings were allowed to curl back around the stigma. In this process, pollen could be dislodged. In addition, the release of sperm cells from germinated pollen tubes could be earlier than ovule maturation. It has been shown that the embryo sac becomes mature as pollen grains reach maturity (Xi, 1991). Discordance of pollen germination and ovule maturation could also reduce the chance of success. For the concurrent pollination to be more successful, the operation time needs to be later in the night allowing for the coordination of pollen germination and ovule maturation and enlargement of peanut flower buds. Therefore, under our greenhouse conditions, conventional “separate” pollination schedule is still the method of choice.
Operator effects
Significant differences were found among the three groups of operators in which group 2 had the highest success rate of 22% followed by group 3 and group 1 (Fig. 2B). Groups 1 and 2 consisted of operators with at least two years of experience and group 3 consisted of operators trained for only two weeks. It is likely that the inexperience of group 3 contributed to their lower success. There is a possibility that the less trained operators may place the pollen on the elongated anther filaments instead of the tip of the stigma. Group 1 removed wings to expose the stigma for pollination in the morning, which could have caused the stigma to get too dry without the protection of wings, resulting in failed fertilization. Group 2 retained the wings while pollinating and was the most successful among the three groups.
Flower opening
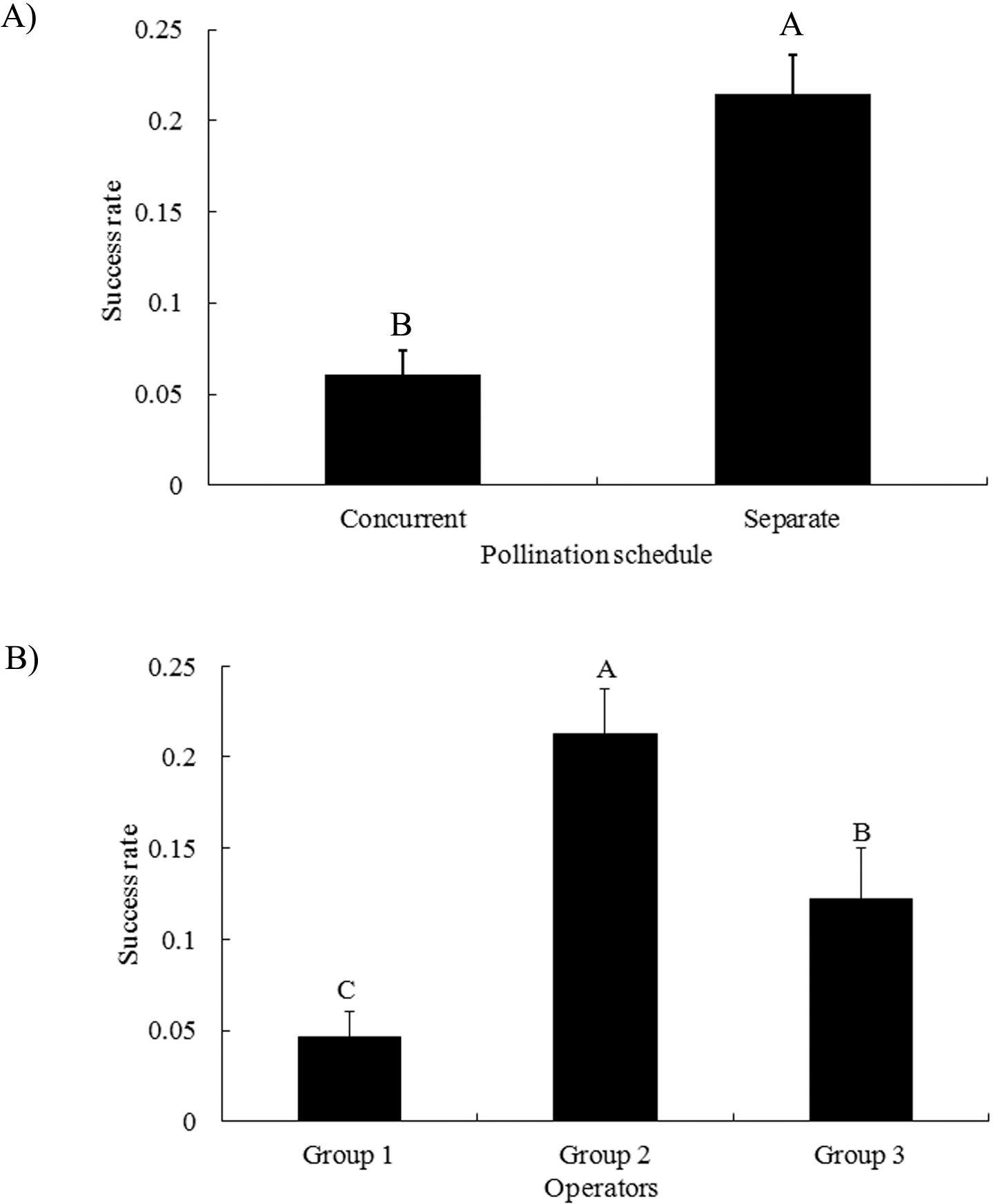
The average success rate of emasculated-full-blooms was higher than, but not significantly different from that of emasculated-but-unopened flowers (Fig. 3A). Hypanthium length of emasculated-full-blooms was similar to that of self-pollinated flowers and significantly longer than that of emasculated-unopened flowers (Fig. 3B). Extension of the hypanthium occurred predominantly after emasculation in our study. Fully opened emasculated flowers had similar hypanthium lengths compared to intact self-pollinated flowers suggesting there was minimum damage to the hypanthium elongation and flower maturation in this group. Association of unopened flowers with reduced hypanthium length suggests some physical damage was done to the female flower buds during emasculation. Although the mean crossing success rate was higher in the full bloom group, no significant statistical difference from the unopened flower group suggests that opening is not the most critical factor contributing to the success of hybridization.
Environment
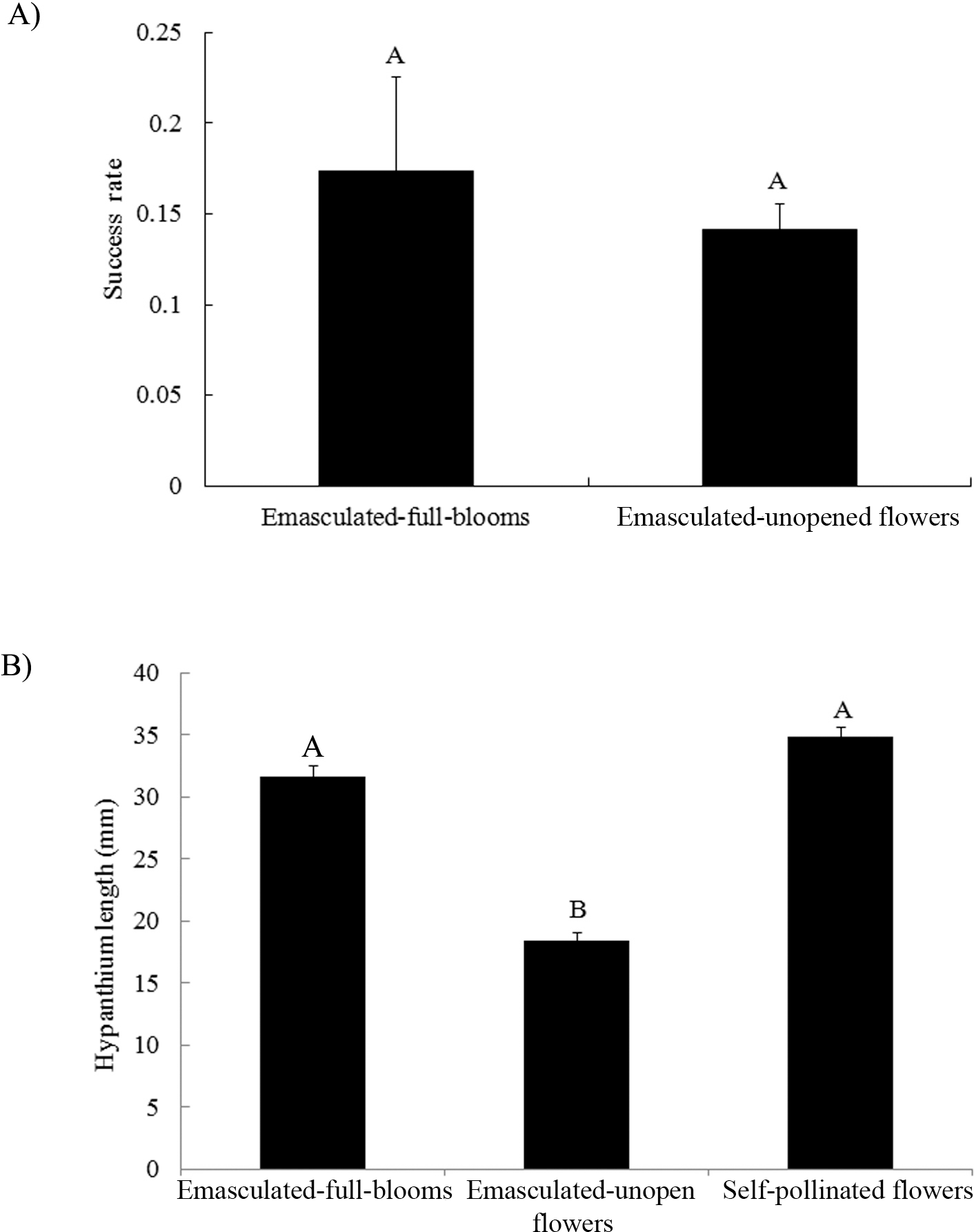
Crossing success rates between parental pairs were also found to be significantly different in which C2244 and C2247 were the lowest in experiment 1 (data not shown). These two least successful crosses had been placed furthest away from the cooling pads in this experiment (Fig. 1A), where average temperature was higher and humidity was lower. Therefore, the low success rate of these two crosses may have been due to the effects of two factors, i.e., the receptiveness of the female genotype and/or environment. In order to separate these two factors, a second experiment was conducted where female genotypes were clustered at either data logger 1 or 2 (Fig. 1B). Significantly higher crossing success was found for females clustered by data logger 1 than those clustered by data logger 2 (Fig. 4A), and there was no significant difference among the genotypes. Since pollination was usually finished by 9:00 hr and fertilization should occur 4-5 hours later (Smith, 1950), average daily temperature and relative humidity were measured between 9:00 and 14:00 hr (Fig. 4B, 4C). Consistently higher temperature and lower humidity were detected by data logger 2 compared to logger 1. Overall average temperatures at logger 1 and 2 were 32.5 C and 36.1 C, respectively. Optimum temperature for pollen germination was reported to be 30 C (Vara Prasad et al., 2011). High temperatures can reduce pollen production and decrease pollen germination rate (Vara Prasad et al., 1999). Both high temperature and low humidity at the center of the greenhouse contributed to the lower success rate of crossing in that location.
Seeds per pod
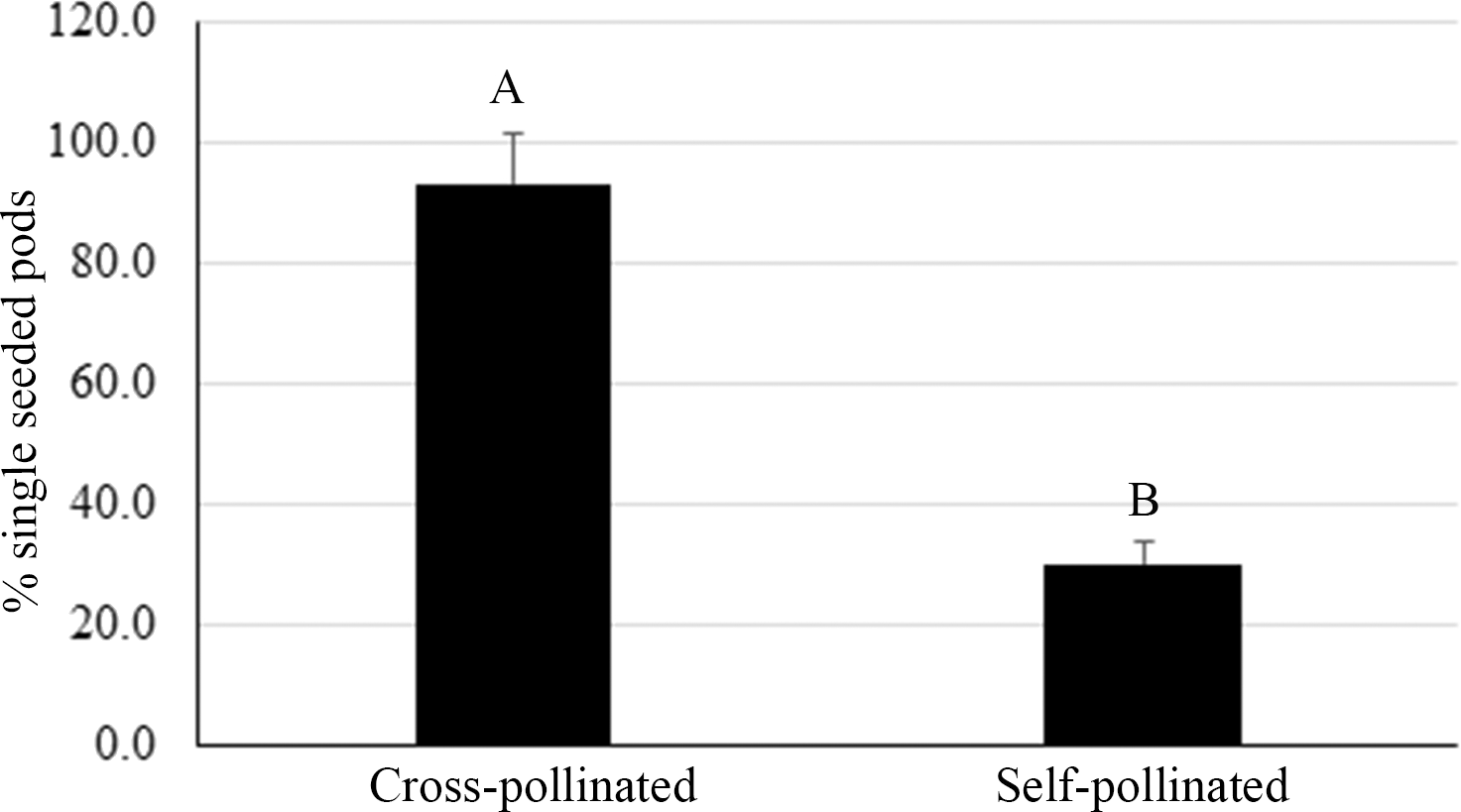
A high frequency of single pods was observed among the hybrids in our breeding program. In the third experiment, all pods were harvested from three crosses whose female plants are runner type, typically producing double pods. After genotyping all seeds for hybridity, it was found that the self-pollination derived pods from these runner-type female plants produced an average of 30% single-seeded pods whereas 80% of pods containing hybrid seed were single-seeded (Fig. 5). The method we used to transfer pollen onto the stigma only allows a small number of pollen grains to lodge on the stigma due to its limited surface area. It has been reported that an average of 56% of pollen grains germinate under optimum conditions in a study with 21 peanut genotypes (Kakani et al., 2002). It is possible that a limited viable pollen supply for fertilization in our study could contribute to the increased rate of single-seeded pods from crosses. An alternative method of pollination, reported to achieve a high crossing success rate, is to cut off the staminal tube from the male flower and apply the whole structure onto the stigma of the female plant (Dr. Charles Simpson, pers. comm.). This method requires more training to master the skill of pollination and one male flower can only fertilize one female flower.
In the present study, multiple factors including operator, crossing schedule, temperature and humidity were found to significantly affect the success of artificial hybridization in peanut. The conventional breeding schedule is suitable for our breeding program using a greenhouse facility. Decreased temperature and increased humidity, as well as proper training of operators, are likely to improve the success rate of crossing. Watering the benches and floor immediately after pollination improves the crossing condition and is routinely practiced in our program. Improvement of greenhouse management should reduce hybrid loss due to fungal disease and water logging. Given the already low yield and labor intensiveness of peanut crossing, optimizing the factors presented in this study will improve the efficiency of crossing programs.
Acknowledgements
We acknowledge funding from the Cultivar Development Research Program of the University of Georgia Research Foundation. We greatly appreciate Betty Tylor, Shannon Atkinson, and Rattan Gill for their participation of this project.
Literature Cited
D. J Banks, (1976). Hybridization of peanuts in growth chambers. Peanut Sci 3 : 66 – 69 .
Bertioli, D. J., S. B Cannon, L. Froenicke, G. Huang, A. D Farmer, E. K. Cannon, X. Liu, D. Gao, J. Clevenger, S. Dash, L. Ren, M.C. Moretzsohn, K. Shirasawa, W. Huang, B. Vidigal, B. Abernathy, Y. Chu, C.E. Niederhuth, P Umale, A.C. Araujo, A Kozik, K.Do Kim, M.D. Burow, R.K. Varshney, X. Wang, X. Zhang, N. Barkley, P.M. Guimaraes, S. Isobe, B. Guo, B. Liao, H.T. Stalker, R.J. Schmitz, B.E. Scheffler, S.C. Leal-Bertioli, X. Xun, S.A. Jackson, R. Michelmore and P. Ozias-Akins (2016) The genome sequences of Arachis duranensis and Arachis ipaensis , the diploid ancestors of cultivated peanut, Nat Genet 48 : 438 – 446.
W. D Branch, (1996). Registration of ‘Georgia Green' peanut. Crop Sci 36 : 806 .
W. D Branch, (2007). Registration of ‘Georgia-06G' peanut. J. Plant Registrations 1 : 120 .
W. D Branch, (2007). Registration of ‘Georgia Greener' peanut. J. Plant Registrations 1 : 121 .
W. D Branch, and T. B Brenneman (2008). Registration of ‘Georgia-07W' peanut. J. Plant Registrations 2 : 88 – 91 .
W. D Branch, and A Culbreath (2011). Registration of ‘Georgia-10T' peanut. J. Plant Registrations 5 : 279 – 281 .
Y., Chu, C. L Wu, C. C Holbrook, B. L Tillman, G Person and P Ozias-Akins (2011). Marker-assisted selection to pyramid nematode resistance and the high oleic trait in peanut. The Plant Genome 4 : 110 – 117 .
T. W., Culp, W. K Bailey and R. O Hammons (1968). Natural hybridization of peanuts, Arachis hypogaea L., in Virginia. Crop Sci 8 : 109 – 111 .
D. F Faucette, and D. A Emery (1974). “Preliminary evaluations of techniques for collecting and culturing peanut ( Arachis hypogaea L.) pollen. Peanut Sci 1 : 14 – 18 .
A. P., Favero, C. E Simpson, J. F. M Valls and N. A Vello (2006). Study of the evolution of cultivated peanut through crossability studies among Arachis ipaensis , A. duranensis, and A. hypogaea . Crop Sci 46 : 1564 – 1552 .
D. W Gorbet, and F. M Shokes (2002). Registration of ‘C-99R' peanut. Crop Sci 42 : 2207 .
D. W Gorbet, and B. L Tillman (2008). Registration of ‘Florida-07’ peanut. J. Plant Registrations 3 : 14 – 18 .
D. W Gorbet, and B Tillman (2011). Registration of ‘York’ peanut. J. Plant Registrations 5 : 289 – 294 .
Hammons, R. O 1964 Pedigreed Natural crossing—a new genetic technique Proceedings, Third National Peanut Research Conference, Auburn, Alabama .
G. L Hildebrand, (1974). A note on artificial hybridization of the groundnut ( Arachis hypogaea L.). Rhod. J. Agric. Res 12 : 195 – 196 .
Holbrook, C. C., T. B Brenneman, H. T Stalker, W. C Johnson III, P Ozias-Akins, Y Chu, G Vellidis and D McClusky 2014 Peanut 33 : 173 – 194 In: S Smith, B Diers, J Specht and B Carver (eds) Yield Gains in Major U.S. Field Crops Madison, WI.
C. C Holbrook, and A Culbreath (2008). Registration of ‘Georganic' peanut. J. Plant Registrations 2 : 17 .
C.C., Holbrook, P Timper, A.K Culbreath and C.K Kvien (2008). Registration of ‘Tifguard’ peanut. J. Plant Registrations 2 : 92 – 94 .
T. G., Isleib, C. C Holbrook and D. W Gorbet (2001). Use of plant introduction in peanut cultivar development. Peanut Sci 28 : 96 – 113 .
V. G., Kakani, P. V., Vara Prasad, P. Q Craufurd, T. R Wheeler (2002). Response of in vitro pollen germination and pollen tube growth of groundnut ( Arachis hypogaea L.) genotypes to temperature. Plant, Cell and Environment 25 : 1651 – 1661 .
D. A., Knauft, A. J Chiyembekeza and D. W Gorbet (1992). Possible reproductive factors contributing to outcrossing in peanut ( Arachis hypogaea L.). Peanut Sci 19 : 29 – 31 .
T. A., Lee, D. L Ketring and R. D Powell (1972). Flowering and growth response of peanut plants ( Arachis hypogaea L. var. Starr) at two levels of relative humidity. Plant Physiol 49 : 190 – 193 .
National Agricultural Statistics Service U.S. Dept. of Agric Published Estimates Database NASS-USDA, Washington, DC. http://www.nass.usda.gov/Statistics_by_Subject/Environmental/index.asp accessed October 3, 2016 .
Norden, A. J 1980 Peanut pp 443 – 456 In: W. R Fehr and H. H Hadley (eds.) Hybridization of Crop Plants American Society of Agronomy-Crop Science Society of America , Madison, WI.
A. J Norden, and V. A Rodriguez (1971). Artifical hybridization of peanuts. Oleagineux 26 : 159 – 162 .
M. K., Pandey, E Monyo, P Ozias-Akins, X Liang, P Guimaraes, S. N Nigam, H. D Upadhyaya, P Janila, X Zhang, B Guo, D. R Cook, D. J Bertioli, R Michelmore and R. K Varshney (2012). Advances in Arachis genomics for peanut improvement. Biotechnol. Adv 30 : 639 – 651 .
C. E Simpson, and J. L Starr (2001). Registration of ‘COAN’ peanut. Crop Sci 41 : 918 – 918 .
B. W Smith, (1950). Arachis hypogaea aerial flower and subterranean fruit. Am. J. Botany 37 : 802 – 815 .
E. G., Stone, W. K Bailey, and J. E Bear (1973). Natural outcrossing of peanuts, Arachis hypogaea L., in Puerto Rico. Proc. J. Am. Peanut Res. Edu. Assoc. Inc 5 : 134 – 140 .
R., Swanson, A. F Edlund and D Preuss (2004). Species specificity in pollen-pistil interactions. Annu. Rev. Genet 38 : 793 – 818 .
S. P., Tallury, T. G Isleib, S. C Copeland, P Rosas-Anderson, M Balota, D Singh, and H. T Stalker (2014). Registration of two multiple disease-resistant peanut germplasm lines derived from Arachis cardenasii . Krapov . & W.C Gregory, GKP 10017. J. of Plant Registrations 8 : 86 – 89 .
B Tillman, and D.W Gorbet (2015). Registration of ‘FloRun ‘107’’ peanut. J. of Plant Registrations 9 : 162 – 167
P. V., Vara Prasad, K. J Boote and L. H Allen (2011). Longevity and temperature response of pollen as affected by elevated growth temperature and carbon dioxide in peanut and grain sorghum. Env. Exp. Bot 70 : 51 – 57 .
P. V., Vara Prasad, P. Q Craufurd, V. G Kakani, T. R Wheeler and K. J Boote (2001). Influence of high temperature during pre-and post-anthesis stages of floral development on fruit-set and pollen germination in peanut. Aust. J. Plant Physiol 28 : 233 – 240 .
P. V., Vara Prasad, P. Q Craufurd, and R. J Summerfield (1999). Sensitivity of peanut to timing of heat stress during reproductive development. Crop Sci 39 : 1352 – 1357 .
P. V., Vara Prasad, P. Q Craufurd and T. R Wheeler (2002). Response of in vitro pollen germination and pollen tube growth of groundnut ( Arachis hypogaea L.) genotypes to temperature. Plant, Cell and Environment 25 : 1651 – 1661 .
X Xi, (1991). Development and structure of pollen and embryo sac in peanut ( Arachis hypogaea L.). Bot. Gaz 152 : 164 – 172 .
Z., Xin, J. P Velten, M. J Oliver and J. J Burke (2003). High-throughput DNA extraction method suitable for PCR. Biotechniques 34 : 820 – 826 .
Notes
- First, second, and fourth authors: Research Professional, Technician, and Professor, Department of Horticulture and NESPAL, University of Georgia Tifton Campus, Tifton, GA 31793; Third author: Research Geneticist, USDA-ARS, P.O. Box 748, Tifton, GA 31793. [^]
- Current address of C. L. Wu: Center for Applied Genetic Technologies, 111 Riverbend Rd, Athens, GA 30602 [^] *Corresponding author's E-mail: pozias@uga.edu
Author Affiliations