Introduction
Genomics, the science of analyzing structure and function of the complete set of DNA in an organism, has been applied to plants since sequencing of the Arabidopsis (Arabidopsis thaliana (L.) Heynh.) genome was initiated (Kaul et al., 2000). Although a weed, Arabidopsis was a logical first candidate for DNA sequencing given its small genome size (125 Mb) and use as a genetic model due to small plant stature, rapid life cycle, and rich genetic resources (Ecker, 1998). It also is a member of a plant family, Brassicaceae, which contains important crop plants, namely rapeseed (Brassica napus L.) and cole crops. For Arabidopsis, research long ago entered the post-genomics era of transcriptomics, proteomics and metabolomics. The first crop plant genome to be sequenced was rice (Oryza sativa L.) because of its global importance and relatively small genome size (420 Mb) (Goff et al., 2002; Yu et al., 2002). With advances in sequencing technology and decreases in the cost per base pair of sequence (Zhao and Grant, 2011), genomics for crop species with larger genomes has become accessible. Most notably, the cereal crops maize (Zea mays L.) and barley (Hordeum vulgare L.), with genome sizes of 2.3 and 5.1 Gb, respectively, and the legume crop soybean (Glycine max (L.) Merr.) (1.1 Gb) have now been sequenced (Mayer et al., 2012; Schmutz et al., 2010; Schnable et al., 2009), as have several model and minor legumes (Cannon et al., 2009; Varshney et al., 2012; Varshney et al., 2013). At last, peanut, Arachis hypogaea L., is on the radar scope for whole genome sequencing, through an international effort coordinated by the Peanut Foundation and substantially funded by the U.S. peanut industry (Peanutbioscience, 2012). The history of this effort can be found in Guo et al. (2013).
The cultivated peanut genome will be challenging to assemble because of its size (2.8 Gb) and complexity (polyploidy). Peanut is an allotetraploid derived from the hybridization of two progenitor diploid species, most likely A. duranensis Krapov. & W.C.Greg. and A. ipaensis Krapov. & W.C.Greg. (Kochert et al., 1996). The tetraploidization event imposed a domestication bottleneck thereby constraining genetic diversity within cultivated peanut. Furthermore, the two progenitor genomes are diverged from one another by only 3–3.5 million years (Moretzsohn et al., 2013; Nielen et al., 2012); therefore, their DNA sequences have high levels of similarity, adding to the difficulty for assembly of a tetraploid sequence that is essentially a merger of the two diploid genomes. In anticipation of the assembly difficulty, the two diploid progenitor genomes are also being sequenced.
With the 2012 launch of genome sequencing of the reference genotype ‘Tifrunner’ (Holbrook and Culbreath, 2007) and the two progenitor diploids, APRES became a preferred venue to describe to the larger peanut community the broader constellation of research and advancements expected in the near future through a symposium entitled, “The orphan legume genome whose time has come”.
The assembly challenges for the peanut genome were illustrated by Scott Jackson (Jackson, 2012), University of Georgia, through an analogy to variants of Vincent Van Gogh's “Sunflowers”. While advances in sequencing technologies have made DNA sequencing more affordable, they also have been based on output of shorter sequences which are more difficult to computationally piece back together in proper order. Imperfect reconstruction of the tetraploid peanut genome is expected when the two subgenomes are so similar to one another that many sequence regions from each will collapse into one (Jackson et al., 2011). While many other crops also are polyploid, their subgenomes are more evolutionarily distant than those of peanut, e.g., cotton (7–8 million years ago) and soybean (13 million years ago) (Jackson and Chen, 2010; Schmutz et al., 2010), and are thus more easily parsed during genome assembly (Schlueter et al., 2007). The prediction is that assemblies of the diploid Arachis progenitor genome sequences will guide assembly of the tetraploid peanut genome sequence. To this end, Lutz Froenicke, UC Davis, provided an update on sequencing of the gene space of A. duranensis and A. ipaensis (Froenicke, 2012).
What is the anticipated impact of a peanut genome sequence on the peanut industry? The peanut industry encompasses growers, shellers, and manufacturers who provide products for consumers. The U.S. peanut industry can claim the highest productivity, highest quality peanut product in the world (FAOSTAT, 2013; American Peanut Council, 2011). In order to maintain that claim while remaining economically competitive, the industry advocates increased yields with concurrent decreased production costs that can be attained in part through genetic improvement and management of pests and diseases with host plant resistance (Valentine, 2012). Nutritional and processing quality must likewise be maintained as productivity increases. Peanut cultivar improvement through breeding began in the early part of the 20th century, and as a result, yield gains due to genetic gain have been impressive, yet gains have not kept pace with corn, cotton, and soybean. Through collective improvements in genetics and cultural practices, average U.S. production increased from 964 kg/ha in 1940 to 4695 kg/ha in 2012 (Holbrook et al., 2013a). A peanut genome sequence will enrich the set of molecular markers that can be applied toward more rapid genetic selection for multiple target traits including yield, biotic and abiotic stress tolerance, and seed quality. For example, the soybean genome sequence led to the discovery of more than 50,000 genetic markers whereas marker numbers had remained in the hundreds prior to sequencing. While U.S. breeders are focused primarily on traits of importance to U.S. markets that affect the economics of production (the estimated annual loss in revenue due to pests and diseases in Georgia alone is more than $100 million (Williams-Woodward 2013)), enriching molecular tools for peanut breeding will have global impact on peanut improvement, leading to a healthier and even more nutritious, protein- and calorie-rich product to help alleviate malnutrition in developing countries.
In order to progress from genome sequence to translational genomics, sequence variation must be associated with specific traits of interest. With advances in sequencing technologies, it is now quicker to generate a genome sequence than to develop and analyze populations segregating for those traits of interest. Corley Holbrook summarizes the current status of U.S. efforts to generate mapping populations segregating for multiple disease resistances as well as productivity and quality traits (Holbrook et al., 2012, 2013b). Reliable phenotyping of populations is essential for association of markers with traits, and phenotyping requires replicated testing over multiple years and ultimately in multiple environments. Therefore, the time frame and labor intensiveness of phenotyping emerges as the bottleneck for translational genomics. The Peanut Genome Project has incorporated a significant component for phenotyping (Table 1, component 5), recognizing that the value of genome sequence will be realized by associating sequences with phenotypes. Some of this value already has been extracted through smaller-scale genetic mapping projects. Baozhu Guo (Guo et al., 2012, 2013) presents an update on genetic mapping of recombinant inbred line (RIL) populations of peanut which illustrates the advances made in marker development, from restriction fragment length polymorphisms (RFLPs) and random amplified polymorphic DNAs (RAPDs) in the 1990s to expressed sequence tag-simple sequence repeats (EST-SSRs) in recent years. Even with the now thousands of molecular markers available for cultivated peanut, the polymorphism information content for most is very low and often only 5–10% of markers screened are polymorphic between any one parental pair. Adopting single nucleotide polymorphisms (SNPs) as markers and expanding marker discovery by whole genome sequencing and resequencing will catapult peanut translational genomics tools to parity with major crops.
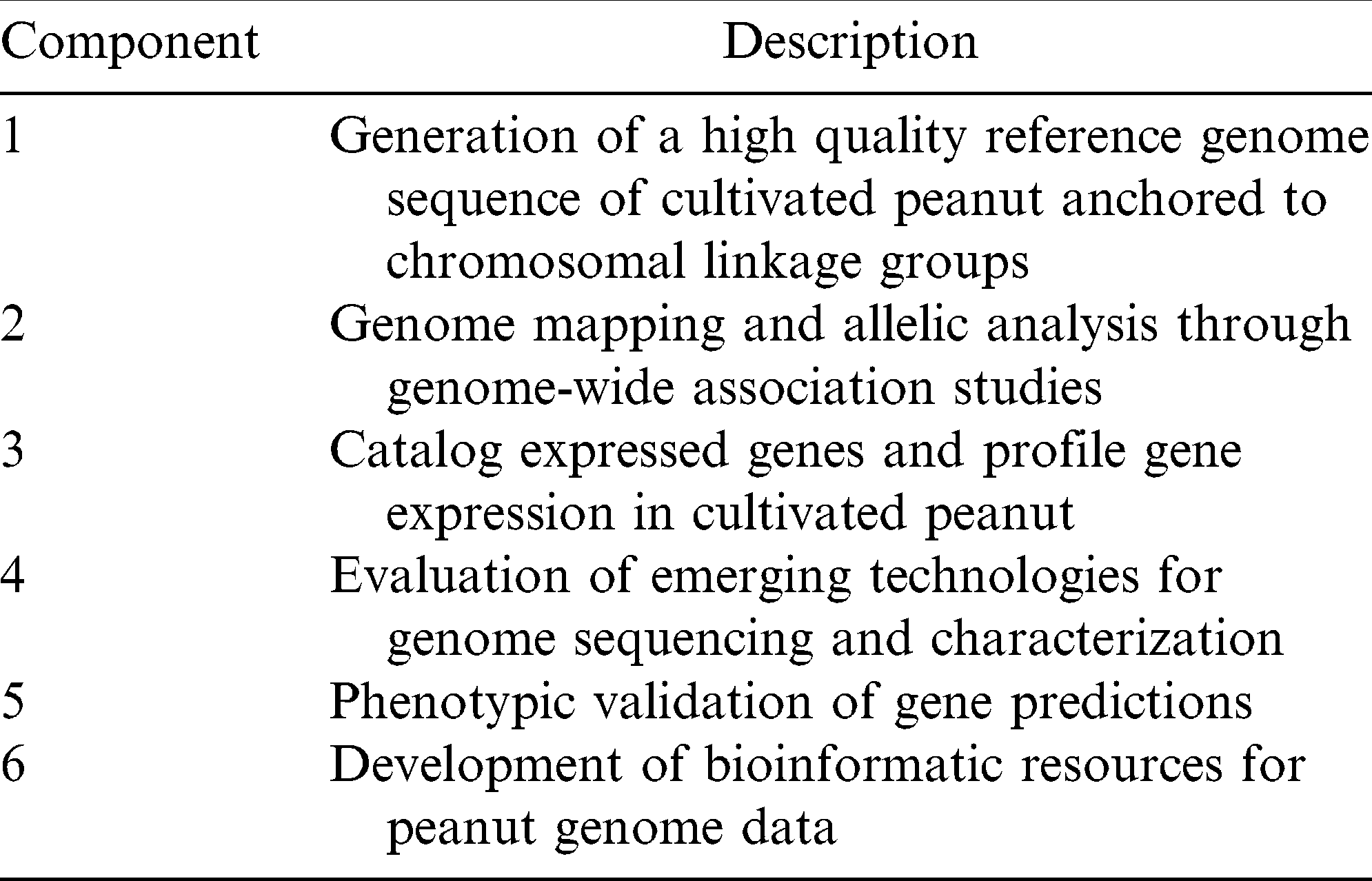
Components of the Peanut Genome Project Action Plan (www.peanutbioscience.com).
One essential component to facilitate molecular breeding is a toolbox enabling access to genomic and phenotypic information along with marker associations that can be easily implemented for marker-assisted breeding. Steven Cannon, USDA-ARS, presented a vision for peanut modelled on existing informatics platforms for other legumes such as SoyBase (soybase.org) and the Legume Information System (LIS; www.comparative-legumes.org) (Cannon 2012). The latter was built with the knowledge, initially from genetic maps, that a high degree of synteny or colinearity (gene position and order) often exists between orthologous chromosome regions of closely related species. Thus, comparative genomics can be exploited to search for candidate genes underlying a specific phenotype that may occur across species or to predict gene function (Young and Bharti, 2012). The web portal, PeanutBase.org, now is online with minimal content but is expected to grow rapidly as the peanut genome project matures.
Finally, the breadth of the peanut genome project to encompass wild diploid as well as cultivated tetraploid species, paralleled by renewed interest in utilizing diploid genetic resources to mine for allelic diversity (Bertioli et al., 2011; Stalker et al. 2012, 2013), will promote the use of wild genetic resources in breeding through introgression pathways (Favero et al., 2006; Simpson, 2001). Tom Stalker, North Carolina State University, reviewed the status of wild-species germplasm collections, resistance attributes, taxonomic relationships, crossability, and molecular variation (Stalker et al. 2012, 2013). Molecular variation between cultivated and wild species is much greater than between accessions within the cultivated species; therefore, genome sequence will obtain highly effective application for monitoring introgression of wild chromosome segments in breeding (Chu et al., 2011; Nagy et al., 2010).
Detailed papers originating from three speakers in the Symposium are published in this issue and recap the utilization of wild Arachis species in breeding (Stalker et al. 2013), population development in cultivated peanut (Holbrook et al. 2013b) and the history of the Peanut Genome Project along with the status of genetic mapping in peanut (Guo et al. 2013). Peanut now has “graduated” from a position of orphan crop to one that soon will be replete with genetic and genomic resources (Varshney et al. 2013).
Literature Cited
American Peanut Council 2011 U.S. quality control and research http:// www.peanutsusa.com (verified August 28, 2013) .
Bertioli D.J Seijo G Freitas F.O Valls J.F.M Leal-Bertioli S.C.M and Moretzsohn M.C 2011 An overview of peanut and its wild relatives Plant Genetic Resources-Characterization and Utilization 9 : 134 – 149 .
Cannon S.B May G.D and Jackson S.A 2009 Three sequenced legume genomes and many crop species: Rich opportunities for translational genomics Plant Physiol. 151 : 970 – 977 .
Cannon S.B 2012 Bioinformatics resources for crop improvement in peanut and across the legumes Proc. Amer. Peanut Res. Educ. Soc. 44 : 57 .
Chu Y Wu C.L Holbrook C.C Tillman B.L Person G and Ozias-Akins P 2011 Marker-assisted selection to pyramid nematode resistance and the high oleic trait in peanut The Plant Genome 4 : 110 – 117 .
Ecker J.R 1998 Genome sequencing - genes blossom from a weed Nature 391 : 438 – 439 .
FAOSTAT 2013 Food and Agriculture Organization of the United Nations Crop Production Database http://www.faostat.fao.org (verified August 28, 2013) .
Favero A.P Simpson C.E Valls J.F.M and Vello N.A 2006 Study of the evolution of cultivated peanut through crossability studies among Arachis ipaensis, A. duranensis, and A. hypogaea Crop Sci. 46 : 1546 – 1552 .
Froenicke L Beitel C Scaglione D Michelmore R.W Bertioli D Moretzsohn M.C Guimaraes P Leal-Bertioli S.C.M Pandey M Upadhyaya H.D and Varshney R.K 2012 Generation of ultra-dense genetic maps for the A and B genomes of peanut Proc. Amer. Peanut Res. Educ. Soc. 44 : 58 – 59 .
Goff S.A Ricke D Lan T.H Presting G Wang R.L et al 2002 A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science 296 : 92 – 100 .
Guo B.Z Pandey M.K Culbreath A.K and Varshney R.K 2012 Recent advances in molecular genetic linkage maps of cultivated peanut (Arachis hypogaea L.) Proc. Amer. Peanut Res. Educ. Soc. 44 : 58 .
Guo B Pandey M He G Zhang X Liao B Culbreath A Varshney R Nwosu V Wilson R and Stalker T Recent advances in molecular genetic linkage maps of cultivated peanut Peanut Sci. 40 : In press .
Holbrook C.C and Culbreath A.K 2007 Registration of ‘Tifrunner’ peanut J. Plant Registr. 1 : 124 .
Holbrook C.C Isleib T.G Ozias-Akins P Chu Y Knapp S.J Tillman B Wu C.L Guo B Gill R and Burow M.D 2012 Development and phenotyping of recombinant inbred line (RIL) populations Proc. Amer. Peanut Res. Educ. Soc. 44 : 56 – 57 .
Holbrook C.C Brenneman T.B Stalker H.T Johnson W.C Ozias-Akins P Chu Y Vellidis G and McClusky D 2013a Yield advances in peanut In: Specht J Diers B Carver B and Smith S (eds.) Yield Gains in Major U.S. Field Crops CSSA , Madison, WI , (In press) .
Holbrook C Isleib T Ozias-Akins P Chu Y Knapp S Tillman B Guo B and Gill R 2013b Development and phenotyping of recombinant inbred line (RIL) populations for peanut Peanut Sci. 40 In press .
Jackson S and Chen Z.J 2010 Genomic and expression plasticity of polyploidy Curr. Opin. Plant Biol. 13 : 153 – 159 .
Jackson S.A Iwata A Lee S.H Schmutz J and Shoemaker R 2011 Sequencing crop genomes: Approaches and applications New Phytol. 191 : 915 – 925 .
Jackson S.A 2012 Genome sequences in Arachis Proc. Amer. Peanut Res. Educ. Soc. 44 : 56 .
Kaul S Koo H.L Jenkins J Rizzo M Rooney T et al 2000 Analysis of the genome sequence of the flowering plant Arabidopsis thaliana Nature 408 : 796 – 815 .
Kochert G Stalker H.T Gimenes M Galgaro L Lopes C.R and Moore K 1996 RFLP and cytogenetic evidence on the origin and evolution of allotetraploid domesticated peanut, Arachis hypogaea (Leguminosae) Amer. J. Bot. 83 : 1282 – 1291 .
Mayer K.F.X Waugh R Langridge P Close T.J Wise R.P and Intl. Barley Genome Sequencing Consortium 2012 A physical, genetic and functional sequence assembly of the barley genome Nature 491 : 711 – 716 .
Moretzsohn M.C Gouvea E.G Inglis P.W Leal-Bertioli S.C.M Valls J.F.M and Bertioli D.J 2013 A study of the relationships of cultivated peanut (Arachis hypogaea) and its most closely related wild species using intron sequences and microsatellite markers Ann. Bot. 111 : 113 – 126 .
Nagy E.D Chu Y Guo Y.F Khanal S Tang S.X Li Y Dong W.B.B Timper P Taylor C Ozias-Akins P Holbrook C.C Beilinson V Nielsen N.C Stalker H.T and Knapp S.J 2010 Recombination is suppressed in an alien introgression in peanut harboring Rma, a dominant root-knot nematode resistance gene Mol. Breed. 26 : 357 – 370 .
Nielen S Vidigal B.S Leal-Bertioli S.C.M Ratnaparkhe M Paterson A.H Garsmeur O D'hont A Guimaraes P.M and Bertioli D.J 2012 Matita, a new retroelement from peanut: Characterization and evolutionary context in the light of the Arachis A-B genome divergence Mol. Genet. Genom. 287 : 21 – 38 .
Peanutbioscience 2013 The peanut genome project, http://peanutbioscience.com/peanutgenomeproject.html (verified August 28, 2013) .
Schlueter J.A Lin J.Y Schlueter S.D Vasylenko-Sanders I.F Deshpande S Yi J O'bleness M Roe B.A Nelson R.T Scheffler B.E Jackson S.A and Shoemaker R.C 2007 Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing BMC Genomics 8 : 330 .
Schmutz J Cannon S.B Schlueter J Ma J.X Mitros T et al 2010 Genome sequence of the palaeopolyploid soybean Nature 465 : 178 – 183 .
Schnable P.S Ware D Fulton R.S Stein J.C Wei F et al 2009 The B73 maize genome: Complexity, diversity, and dynamics Science 326 : 1112 – 1115 .
Simpson C.E 2001 Use of wild Arachis species/introgression of genes into A. hypogaea L Peanut Sci. 28 : 114 – 116 .
Stalker H.T Tallury S.P Ozias-Akins P Bertioli D.A and Leal-Bertioli S.C.M 2012 The value of diploid peanut relatives for breeding and genomics Proc. Amer. Peanut Res. Educ. Soc. 44 : 57 – 58 .
Stalker H.T Tallury S.P Ozias-Akins P Bertioli D and Leal Bertioli S.C 2013 . The value of diploid peanut relatives for breeding and genomics Peanut Sci. 40 : (In press)
Valentine H 2012 Potential economic impact of the peanut genomic project (PGP) Proc. Amer. Peanut Res. Educ. Soc. 44 : 56 .
Varshney R.K Chen W Li Y Bharti A.K Saxena R.K et al 2012 Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers Nature Biotech. 30 : 83 – 89 .
Varshney R.K Song C Saxena R.K Azam S Yu S et al 2013 Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement Nature Biotech. 31 : 240 – 246 .
Varshney R.K Mohan S.M Gaur P.M Gangarao N.V.P.R Pandey M.K et al 2013 Achievements and prospects of genomics-assisted breeding in three legume crops of the semi-arid tropics, Biotech. Adv. http://dx.doi.org/10.1016/j.biotechadv.2013.01.001 .
Williams-Woodward J 2013 Georgia plant disease loss estimates 2011 University of Georgia Coop. Exten. Annu. Publ. 102-4.
Young N.D and Bharti A.K 2012 Genome-enabled insights into legume biology Annu. Rev. Plant Biol. 63 : 283 – 305 .
Yu J Hu S.N Wang J Wong G.K.S Li S.G et al 2002 A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science 296 : 79 – 92 .
Zhao J and Grant S.F.A 2011 Advances in whole genome sequencing technology Curr. Pharm. Biotech. 12 : 293 – 305 .
Notes
- Department of Horticulture, NESPAL, and Institute of Plant Breeding, Genetics and Genomics, The University of Georgia, Tifton, Georgia 31793 * Corresponding author’s E-mail: pozias@uga.edu
Author Affiliations