Introduction
Peanut (Arachis hypogaea L.) is one of the world's most important commercial crops, cultivated in over 100 tropical and subtropical countries (http://faostat.fao.org/default.aspx, accessed July 2008). Peanut products are widely consumed because of their excellent nutritional content and flavor. Peanut production and seed quality, however, can be severely affected by fungal and viral diseases (Melouk and Shokes, 1995). Aflatoxin contamination of peanuts is an industry-wide problem affecting peanut growers, manufacturers and consumers (Jelinek et al. 1989). Shelled peanuts having greater than 20 ppb (parts per billion) of aflatoxin, a known human carcinogen, exceed the Food and Drug Administration action level for foods other than milk (www.cfsan.fda.gov/˜lrd/fdaact.html). In peanut, aflatoxin is produced by Aspergillus flavus and A. parasiticus (Wilson, 1995). From 1993 to 1996 aflatoxin outbreaks in top peanut-producing states of the USA caused average annual losses of $26 million to its southeastern peanut industry (Lamb and Sternitzke, 2001). Although peanut germplasm with reduced preharvest aflatoxin contamination is being screened in breeding populations (Holbrook et al. 2000), peanut growers currently can take no direct action to control A. flavus and A. parasiticus, other than irrigation. Therefore, genetic engineering of peanut with antifungal genes is an attractive prospective way to reduce pre-harvest aflatoxin contamination.
Transformation of peanut has been accomplished by several different methods, including particle bombardment (Higgins et al. 2004; Livingstone and Birch, 1999; Magbanua et al. 2000; Ozias-Akins et al. 1993; Yang et al. 1998), Agrobacterium-mediated transformation (Cheng et al. 1997; Rohini and Rao, 2001; Sharma and Anjaiah, 2000), and electroporation (De Padua et al. 2000). Individual genes that confer agronomic traits have been integrated into the peanut genome such as bialophos resistance (bar) for herbicide tolerance (Brar et al. 1994), Bacillus thuringiensis (Bt) toxin cryIA(c) for insect resistance (Singsit et al. 1997), viral nucleocapsid or coat protein genes for virus resistance (Higgins et al. 2004; Li et al. 1997; Magbanua et al. 2000; Sharma and Anjaiah, 2000; Yang et al. 1998; Yang et al. 2004), chitinase, glucanase, and oxalate oxidase to control fungal diseases (Chenault et al. 2005; Chenault et al. 2002; Livingstone et al. 2005; Rohini and Rao, 2001).
Rajasekaran et al. (2000) reported that the expression of a chloroperoxidase gene from Pseudomonas pyrrocinia (cpo-p) (Wiesner et al. 1988; Wolfframm et al. 1993) in transgenic tobacco resulted in significant inhibition of A. flavus hyphal growth and reduced leaf anthracnose lesions caused by Colletotrichum destructivum. Although the exact mechanism of antifungal activity in cpo-p transgenic tobacco was not determined, it had been previously shown that haloperoxidases (HPO) catalyze the peroxidation of halides to form hypohalites: H2O2 + X− → H2O + −OX, where X− is a non-fluoride halide (van Pée, 1996). The haloperoxidase CPO-P also catalyzes the formation of peracetic acid: AcOH + H2O2 → AcOOH + H2O (Picard et al. 1997; van Pée, 1996). Both hypohalites and peracetic acid are strong antimicrobial agents, and might be responsible for the increase of antifungal activity of CPO-P transgenic tobacco (Jacks et al. 2000; Rajasekaran et al. 2000; van Pée, 1996). Compared with most other HPOs, CPO-P does not require a heme prosthetic group, proteins which usually are not available in plants and are encoded by other genes, making it very suitable for plant transformation.
The resistance of cpo-p transgenic tobacco to A. flavus (Rajasekaran et al. 2000), coupled with the development of a genotype independent peanut transformation system (Ozias-Akins et al. 1993), made CPO-P in transgenic peanut an attractive target to examine the potential to reduce pre-harvest aflatoxin contamination. The objectives of the present study were 1) to insert the cpo-p gene into peanut somatic embryos via particle bombardment, 2) to examine the bacterial cpo-p gene expression in peanut, and 3) to assay its potential antifungal activity to A. flavus.
Materials and Methods
Plant material and in vitro cultures
Embryogenic cultures of peanut were initiated from mature zygotic embryos (McKently, 1991) of the commercial cultivar, Georgia Green. Seeds were sterilized by shaking twice for 20 min each in 20% Clorox (1% w/v NaOCl), followed by three rinses with sterile deionized water. The plumule part of the embryo axis was excised and cultured on embryogenesis medium, which consisted of FN Lite medium (Samoylov et al. 1998) supplemented with 2.4% sucrose, 3 mg/L (12.42 µM) picloram, 1 g/L glutamine (filter-sterilized) and 0.8% agar. The pH was adjusted to 5.8 prior to autoclaving. Embryogenic cultures were grown in the dark at 26 ± 2 °C for up to 12 months and bombarded approximately 2 weeks after subculture under the assumption that the cultures were between lag and stationary phases of growth.
Plasmid constructs
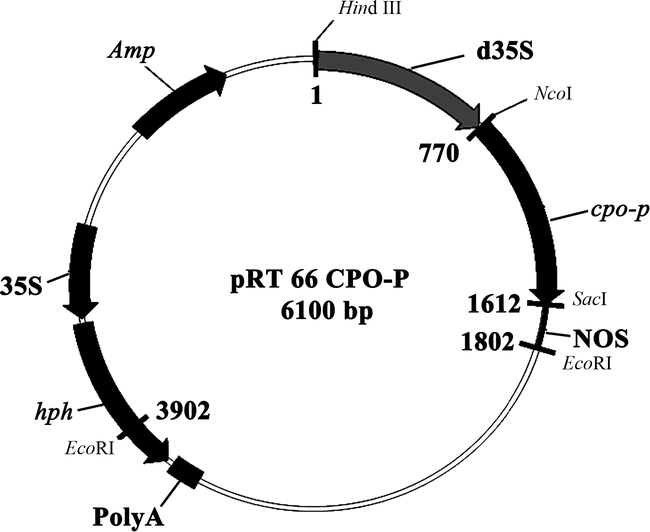
The vector (pBI101) contained the cpo-p gene from P. pyrrocinia (Wolfframm et al. 1993). The cpo-p coding region with a double CaMV 35S promoter and nos terminator (1.8 kb) was excised from the pBI101-derivative with HindIII/EcoRI digestion and subcloned into HindIII/EcoRI-digested pRT66, which contains the hygromycin phosphotransferase (hph) coding region under the control of the CaMV 35S promoter (Topfer et al. 1993). The resulting plasmid containing cpo-p and hph is referred to as pRT66 cpo-p (Fig. 1).
Microprojectile bombardment, selection, and regeneration
Twenty clusters of somatic embryos, with 3–5 embryos per cluster, were arranged in the central 2-cm area of a petri dish. Particle bombardment was carried out using a PDS 1000/helium-driven biolistic device (Bio-Rad Laboratories, Hercules, CA) as described previously (Ozias-Akins et al. 1993; Yang et al. 1998), except that 0.6 µm gold particles were used. Tissues were desiccated by uncovering the plates for 2–3 h prior to bombardment in the laminar flow hood. Selection was initiated in liquid embryogenesis medium containing 10 mg/L hygromycin 3 days post-bombardment in the dark. The medium was changed weekly for the first two subcultures to eliminate loose cell debris and agar medium after which it was changed every two weeks and the level of hygromycin was increased to 20 mg/L. A three-step regeneration protocol was followed to obtain plants from stably transformed somatic embryo cell lines (Ozias-Akins et al. 1993).
PCR and Southern blot analysis of putative transformants
PCR was performed on hygromycin-resistant cell lines or regenerated putative transgenic plants to check for the presence of the cpo-p gene. Genomic DNA was isolated by the CTAB method (Murray and Thompson, 1980). A 582-bp fragment of the open reading frame of cpo-p was amplified using the sense primer S67 (5′-CAG CCC ATC GTC TTC CAT CAT GGC-3′) and an antisense primer A648 (5′-CGA CTT CAG GTC CTC GGT CTG ATC-3′). The PCR amplification was carried out using the following program: 94 °C for 5 min followed by 40 cycles of 94 °C for 1 min, 60 °C for 1 min, 72 °C for 1 min. The same PCR-amplified 582-bp fragment was also used as a probe for Southern and Northern blot hybridization.
For Southern blot analysis, peanut genomic DNA (10 µg) was digested to completion with HindIII or EcoRI and electrophoresed on a 0.8% agarose gel. DNA was transferred to GeneScreen Plus nylon membrane (PerkinElmer, Waltham, MA) using 0.4 N NaOH. The 582-bp cpo-p probe was labeled with α-32P-dCTP using a random priming DNA labeling kit (Roche Applied Science, Indianapolis, IN) and hybridization was carried out overnight at 65 °C in the solution of Church and Gilbert (Church and Gilbert, 1984). Following hybridization, the membrane was washed at 65 °C for 15 min in 25 mL 2× SSPE/1% SDS, 15 min in 100 mL of 0.5× SSPE/1% SDS, and 15 min in 100 mL 0.1× SSPE/1% SDS. Hybridization signal was detected using a Cyclone Imaging System equipped with OptiQuant software (PerkinElmer, Waltham, MA).
Analysis of transgene expression
Total plant RNA was isolated from selected embryogenic callus or young leaves (50–100 mg) using the Qiagen RNeasy Minikit (Qiagen, Valencia, CA). Total RNA (10 µg) was electrophoresed on a 1.0% denaturing formaldehyde (2.2 M) gel using 1× MOPS running buffer as described (Sambrook et al. 1989). RNA was transferred to GeneScreen Plus membrane using 7.5 mM NaOH and hybridized with the PCR-amplified 582-bp 32P-labelled cpo-p gene fragment.
The method for Western blot analysis of CPO-P was modified from Rajasekaran et al. (2000). Leaf tissue was ground to a fine powder in liquid nitrogen and extraction buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 5% 2-mercaptoethanol) was added (0.5 mL per g of leaf). The homogenate was centrifuged twice at 13,000 g at room temperature for 5 min each to remove cell debris. The supernatant was collected and total protein in each sample was determined using the BioRad Protein Assay (Bio-Rad Laboratories, Hercules, CA) with bovine serum albumin as the standard. Aliquots of the resulting supernatants (40 µg crude protein) were denatured by heating with Laemmli buffer at 100 °C for 4 min and separated on a 12% polyacrylamide gel along with protein molecular weight standards (14.4–97.4 kDa Low-Range, Bio-Rad, Hercules, CA), and transferred to PVDF membranes (Bio-Rad Laboratories, Hercules, CA) with an electroblotter (Bio-Rad Laboratories, Hercules, CA). After blocking with 0.5% non-fat dry milk in PBST buffer, the membranes were probed with CPO-P polyclonal antiserum at 1∶1000 dilution and goat anti-rabbit alkaline phosphatase secondary antibody conjugate (Sigma-Aldrich, St. Louis, MO) diluted at 1∶1000. Nitro-blue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate reagents were used for signal detection.
In vitro bioassay of antifungal activity in transgenic tissue extracts
The in vitro analysis of antifungal activity of transgenic peanut extract to A. flavus was based on the method described previously (DeLucca et al. 1997; Rajasekaran et al. 2000). For tissue extract preparation, 1–2 g of embryogenic callus or peanut leaves were frozen in liquid nitrogen and ground without buffer. The sample extract was collected by centrifuging ground tissues at 10,000 g for 10 min at 4 °C. A conidial suspension (105 cells/mL) of A. flavus NRRL3357 was pre-germinated for 8 h in potato dextrose broth (Voight Global Distribution, Lawrence, KS) and 17 µL were added to 153 µL of plant extract and incubated for 1–4 h at 30 °C. Three 50 µL aliquots were taken from each sample and spread onto three potato dextrose agar plates. The number of colony forming units was counted after incubation at 30 °C for 24–36 h. Antifungal assays were repeated at least three times for each transgenic sample and non-transgenic control Georgia Green. One-way ANOVA was used to analyze the effect of transgenic callus or leaf extracts on conidia germination. Mean separation was performed using the method of Tukey.
In situ inoculation of peanut cotyledons with A. flavus 70-GFP
The EGFP (Enhanced Green Fluorescent Protein) gene (Clontech Laboratories, Mountain View, CA) was placed under the control of a constitutively expressed A. nidulans glyceraldehyde phosphate dehydrogenase (gpdA) gene promoter and the A. parasiticus N-myristoyl transferase (nmt-1) gene transcriptional terminator. All of these elements were subcloned into the plasmid vector pBlueScript-SK (Stratagene) to produce the vector gpd-EGFP (Rajasekaran et al. 2008). Plasmid gpd-EGFP was cotransformed with the vector pSL82 harboring the A. parasiticus nitrate reductase (niaD) gene into the niaD mutant of A. flavus 70. One isolate stably expressing high levels of GFP, designated A. flavus 70-GFP, was used in all experiments.
Mature transgenic peanut cotyledons and controls were inoculated with A. flavus 70-GFP to evaluate fungal spread in situ (Rajasekaran et al. 2005, 2008). Cotyledons were soaked overnight in water and sliced (10 mm × 6 mm × 6 mm) to keep the explants uniform. The A. flavus GFP strain was grown for 7 days at 30 °C on malt extract agar medium (Sigma-Aldrich, St. Louis, MO) before assay. Conidia were harvested by flooding a single plate with 9 mL of 0.01% (v/v) sterile Triton X-100 solution and scraping the surface of mycelium with a sterile pipette. Conidia were diluted to 2 × 104 per mL for seed inoculation studies. About 100 conidia (5 µL) were introduced into cotyledon segments through a needle wound of approximately 3–4 mm depth. A minimum of 10 cotyledon segments per plate and three replicate plates were used per transgenic T4. To maintain high humidity, cotyledons were placed on three layers of Whatman filter papers soaked with water and the petri dishes were sealed with two layers of parafilm. After ten days of incubation at 30 °C the cotyledons were examined for GFP fluorescence of mycelium using the Olympus SZH10 GFP-stereomicroscope and the fungal colonization was quantitatively assessed as follows. Cotyledons were ground in liquid nitrogen, and 500 µL of 50 mM phosphate buffer, pH 7.2, was added to 500 mg of powdered cotyledons and mixed well by vortexing. The samples were centrifuged at 10,000 rpm for 10 min in a swinging bucket rotor. One hundred µL aliquots of supernatant from each sample were placed in a 96-well HP Viewplate, and the fluorescence was measured at an excitation wavelength of 485 nm and an emission wavelength of 535 nm using the Perkin-Elmer HTS 7000 fluorometer. Average fluorescence values were obtained for each sample and were subjected to non-parametric ANOVA using the GraphPad Prism software.
Results and Discussion
Selection and regeneration of transformed callus
Most bombarded embryogenic tissues of Georgia Green showed rapid necrosis in medium containing 20 µg/mL hygromycin, and only a few tissues showed growth after ∼ 8 weeks on liquid selection. Tissues that were actively growing on liquid selection medium were transferred to agar medium containing 20 µg/mL hygromycin, and individual pieces were assigned a unique cell line number. Thirteen bombardment experiments with 107 plates yielded 147 hygromycin-resistant cell lines for an average of 1.37 cell lines per plate, which is within the reported peanut transformation efficiency range of 0.8 to 4.6 hygromycin resistant callus lines per bombardment (Higgins et al. 2004; Ozias-Akins et al. 1993; Wang et al. 1998; Yang et al. 1998). It is difficult to get accurate transformation efficiency data based on somatic embryo numbers bombarded since clusters of somatic embryos contain variable numbers, and some cell lines may have originated from the same transformation event but were separated due to agitation during liquid selection. In our estimation, there were 60–100 embryos present in each bombarded plate; therefore, the transformation efficiency for Georgia Green on a per embryo basis ranged from 1.37–2.28%. Plants were regenerated from 75 hygromycin-resistant cell lines and grown in the greenhouse.
Presence and integration of the cpo-p gene in peanut
The presence of the cpo-p gene was verified by PCR. A 582-bp fragment of the cpo-p gene was amplified from all 24 of the hygromycin-resistant cell lines tested but not from untransformed Georgia Green (data not shown). This result indicated that the hygromycin selection system was very effective for recovery of cpo-p transgenic cell lines from Georgia Green using the pRT 66 cpo-p construct.
Out of 78 putative T0 transgenic plants tested, 67 (86%) of them amplified the 582-bp fragment of cpo-p. As expected, hygromycin selection for transgenic peanut was not 100% efficient for recovery of a linked gene. Similar results were previously shown by Yang et al. (1998), who reported that 92% of cell lines PCR-positive for the hygromycin resistance gene also showed PCR amplification of the nucleocapsid protein gene of tomato spotted wilt virus, and Wang et al. (1998) who reported that 92% of hygromycin-resistant and PCR-positive cell lines showed PCR amplification of the vspB promoter-driven reporter gene, β-glucuronidase. It is likely that the non-selected gene linked with hph was disrupted in some instances during plasmid DNA integration.
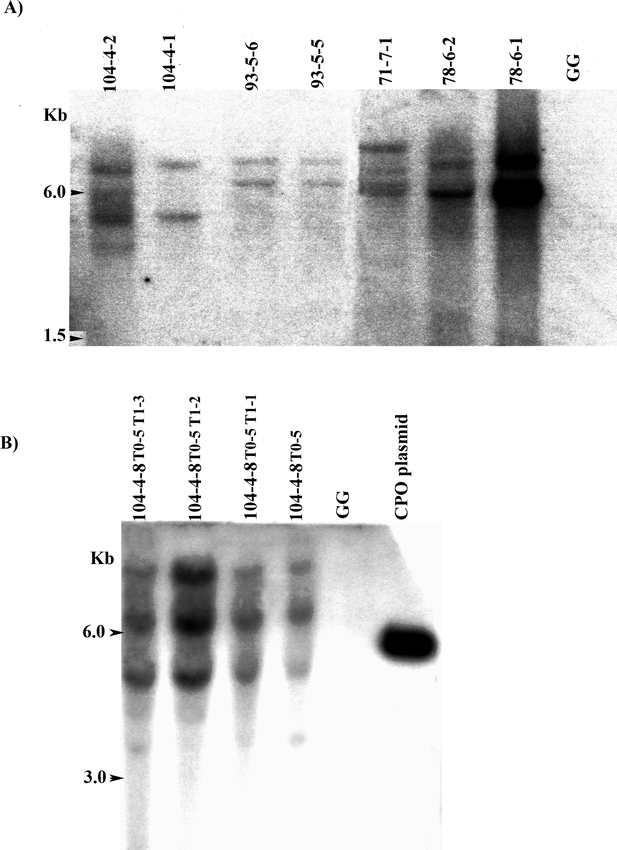
Southern blot analysis with HindIII-digested DNA from hygromycin-resistant cell lines that were also confirmed to be PCR-positive for cpo-p, showed that most had multiple (2–4) hybridizing bands (Fig. 2A). Since the pRT66 cpo-p plasmid has only a single HindIII recognition site, these results indicated that multiple insertions were prevalent in the transgenic material. The particle bombardment method often results in the integration of multiple copies of transgenes (Meyer, 2000), which has frequently been observed in peanut particle bombardment experiments (Higgins et al. 2004; Livingstone and Birch, 1999; Yang et al. 1998). In addition, the band patterns revealed that some cell lines, such as 93-5-5 and 93-5-6 that originated from the same container during liquid selection, represented the same transformation event. Reducing the liquid selection time or replacing it with selection on agar medium could reduce the chance that multiple lines would originate from one event. However, selection on agar is more laborious for large-scale experiments and might increase the chance of recovering escapes. Southern blot analysis with transgenic T0 and T1 progeny plants from line 104-4-8 confirmed stable integration and transmission of the cpo-p gene (Fig. 2B).
Northern and Western blot analysis of cpo-p expression in peanut
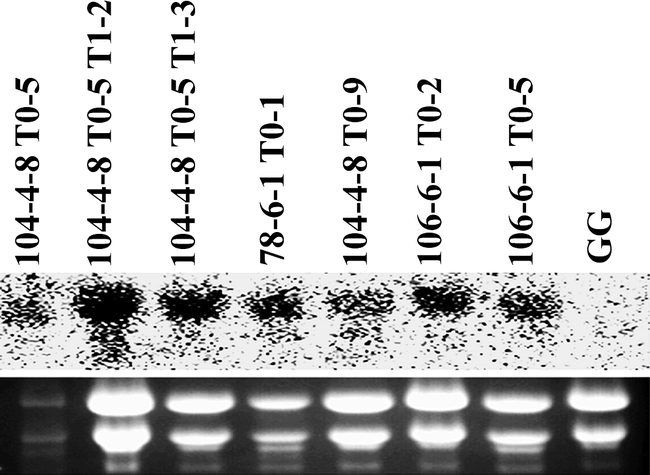
To examine the bacterial cpo-p gene expression in peanut, and to efficiently select useful transgenic cell lines for regeneration, Northern blot analysis was conducted on hygromycin-resistant embryogenic cell lines. Eighteen out of twenty cell lines showed transcripts of cpo-p indicating that a high (90%) percentage of hygromycin-resistant somatic embryos expressed the gene (data not shown). The presence of the cpo-p transcript in transgenic plants regenerated from the cell lines expressing cpo-p was confirmed by Northern blot analysis of total RNA from leaf tissue (Fig. 3).
The cpo-p gene product, a 32 kD protein (Wolfframm et al. 1993), was detected by Western blot analysis in transgenic plants (Fig. 4). The result demonstrated that this bacterial cpo-p gene is efficiently expressed in transgenic peanut. One regenerated line, J6-2-2 T0-3, which was verified by PCR to be an escape, was also included in the Western blot analysis as a negative control and showed no immunoreactive band thus confirming the PCR data. In summary, 18/20 lines tested by Northern blot analysis showed cpo-p transcription, whereas six of those transcribing cpo-p were tested and shown to express CPO-P at the protein level. Only two lines were subjected to all molecular assays plus bioassay due to their vigor (and thus availability of leaf material) or ability to set seed.
In vitro antifungal activity of transgenic peanut extracts
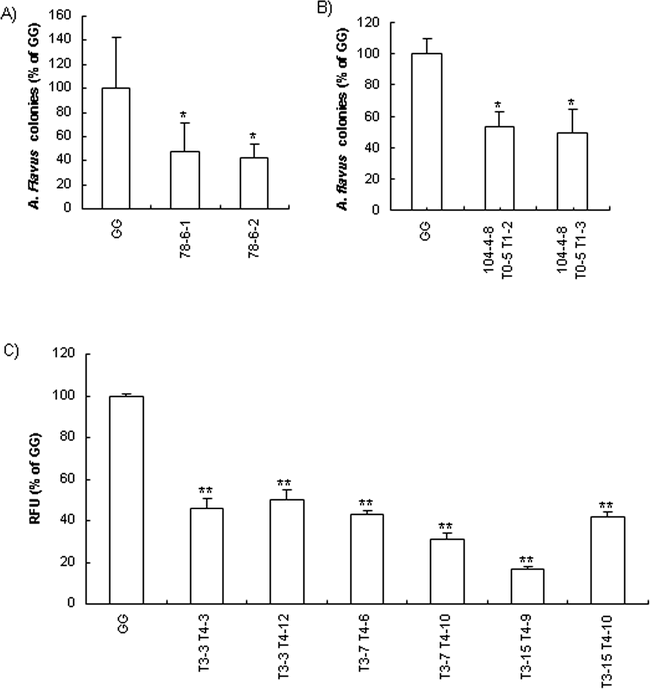
To assay the potential antifungal activity of CPO-expressing transgenic plants was a major objective of this study. Crude protein extracts from embryogenic tissues of two Northern-blot-positive cell lines significantly inhibited (P<0.05) the growth of pre-germinated A. flavus conidia compared with non-transformed Georgia Green (Fig. 5A). Leaf extracts from mature transgenic peanut plants also showed significant antifungal activity for T1 progenies of line 104-4-8 (Fig. 5B). The presence of cpo-p transcript (Fig. 3) and CPO-P protein (Fig. 4) confirmed that the antifungal property of the transgenic progeny plants was due to cpo-p gene expression. Variation in expression and in vitro antifungal activity among individual plants was observed in this study and also was reported in CPO-expressing transgenic tobacco (Jacks et al. 2000; Rajasekaran et al. 2000).
Inhibition of A. flavus spore germination by extracts of embryogenic cultures (A) and leaves (B); inhibition of A. flavus fungal growth on peanut cotyledons from T4 progenies of CPO transgenic line 104-4-8 T0-5 T1-3 T2-3 (C). GG : non-transgenic Georgia Green. * and ** denote significant differences between the transgenic lines and GG at P<0.05 and P<0.01 levels, respectively. Error bars indicate standard deviation. Mean separation was performed by Tukey's method.
Although little data were collected to compare the antifungal activity of extracts from embryogenic tissues with leaf tissues, we did observe that one cell line (71-7-2) with callus extract reduced the number of A. flavus colonies by 50% compared with the control, whereas with leaf extract, the number of A. flavus colonies was reduced by only 20%. Further experiments would be necessary to conclude whether the reduced antifungal activity in leaf extracts might be due to differences in protein accumulation resulting from the physiological or developmental status of the plant or to proteases or other substances that might not be as abundant in callus extracts. Our results do support, however, the use of embryogenic tissue extracts to predict antifungal activity of plant tissue extracts.
In situ inoculation of mature peanut cotyledons with A. flavus 70-GFP
Cotyledon segments from both controls and transgenic groups inoculated with A. flavus 70-GFP strain turned yellow 4–5 days after inoculation due to colonization of cotyledons by the fungus. At the inoculation site the 70-GFP strain showed extensive growth and sporulation. Cotyledons from transgenic seeds showed significantly less (50–80%) fluorescence than the controls indicating a reduction in colonization of transgenic peanut cotyledons by A. flavus 70-GFP strain (Fig. 5C).
Conclusion
The results presented here demonstrate the successful insertion and expression of the cpo-p gene into A. hypogaea cv. Georgia Green using the genotype independent peanut transformation protocol established in our laboratory (Ozias-Akins et al. 1993; Singsit et al. 1997; Wang et al. 1998; Yang et al. 1998). The stable transmission of the cpo-p gene was further confirmed by Southern blot and expression analysis of progeny from one transgenic line. To our knowledge, the bacterial cpo-p gene has been thoroughly studied in only two transgenic plant species, tobacco and cotton, with a vector carrying cpo-p and kanamycin resistance genes (Jacks et al. 2000, 2004; Rajasekaran et al. 2000, 2008). We have previously shown that a codon-modified Bt cryIA(c) gene, which was introduced into three peanut cultivars (Singsit et al. 1997), has potential to provide a barrier to fungal entry by reducing insect damage to peanut tissue. Here, we describe an additional strategy to directly increase the antifungal activity of peanut to the aflatoxin-producing pathogen, A. flavus.
Acknowledgements
The authors thank Thomas E. Cleveland, Jim Gegogeine, Laura Ramos and Yukio Akiyama for helpful discussion. This work was supported by grants from the USDA Multicrop Aflatoxin Elimination Program and The Peanut Foundation. We thank Anne Bell, Evelyn Perry Morgan, and Gregory Ford for technical assistance.
Literature Cited
Brar G. S. , Cohen B. A. , Vick C. L. , and Johnson G. W. 1994 Recovery of transgenic peanut (Arachis hypogaea L.) plants from elite cultivars utilizing ACCELL technology. Plant J 5 : 745 – 753 .
Chenault K. D. , Melouk H. A. , and Payton M. E. 2005 Field reaction to Sclerotinia blight among transgenic peanut lines containing antifungal genes. Crop Sci 45 : 511 – 515 .
Chenault K. D. , Burns J. A. , Melouk H. A. , and Payton M. E. 2002 Hydrolase activity in transgenic peanut. Peanut Sci 29 : 89 – 95 .
Cheng M. , Jarret R. L. , Li Z. , and Demski J. W. 1997 Expression and inheritance of foreign genes in transgenic peanut plants generated by Agrobacterium-mediated transformation. Plant Cell Rep 16 : 541 – 544 .
Church G. M. and Gilbert W. 1984 Genomic sequencing. Proc. Natl. Acad. Sci. USA 81 : 1991 – 1995 .
De Padua V. L. M. , Pestana M. C. , Margis-Pinheiro M. , De Oliveira D. E. , and Mansur E. 2000 Electroporation of intact embryonic leaflets of peanut: Gene transfer and stimulation of regeneration capacity. In Vitro Cell. Dev. Biol. Plant 36 : 374 – 378 .
DeLucca A. J. , Bland J. , Jacks T. J. , Grimm C. , Cleveland T. E. , and Walsh T. J. 1997 Fungicidal activity of cecropin A. Antimicrob. Agents Chemother 41 : 481 – 483 .
Higgins C. M. , Hall R. M. , Mitter N. , Cruickshank A. , and Dietzgen R. 2004 Peanut stripe potyvirus resistance in peanut (Arachis hypogaea L.) plants carrying viral coat protein gene sequences. Transgenic Res 13 : 59 – 67 .
Holbrook C. C. , Kvien C. K. , Rucker K. S. , Wilson D. M. , Hook J. E. , and Matheron M. E. 2000 Preharvest aflatoxin contamination in drought-tolerant and drought-intolerant peanut genotypes. Peanut Sci 27 : 45 – 48 .
Jacks T. J. , De Lucca A. J. , Rajasekaran K. , Stromberg K. , and van Pée K. 2000 Antifungal and peroxidative activities of nonheme chloroperoxidase in relation to transgenic plant protection. J. Agric. Food Chem 48 : 4561 – 4564 .
Jacks T. J. , Cary J. W. , Rajasekaran K. , Cleveland T. E. , and van Pée K-H. 2004 Transformation of plants with a chloroperoxidase gene to enhance disease resistance. U.S. Patent #6,703,540.
Jelinek C. F. , Pohland A. E. , and Wood G. E. 1989 Worldwide occurrence of mycotoxin in food and feeds - an update. J. Assoc. Anal. Chem 72 : 223 – 230 .
Lamb M. C. and Sternitzke D. A. 2001 Cost of aflatoxin to the farmer, buying point, and sheller segments of the Southeast United States peanut industry. Peanut Sci 28 : 59 – 63 .
Li Z. , Jarret R. L. , and Demski J. W. 1997 Engineered resistance to tomato spotted wilt virus in transgenic peanut expressing the viral nucleocapsid gene. Transgen. Res 6 : 297 – 305 .
Livingstone D. M. and Birch R. G. 1999 Efficient transformation and regeneration of diverse cultivars of peanut (Arachis hypogaea L.) by particle bombardment into embryogenic callus produced from mature seeds. Mol. Breed 5 : 43 – 51 .
Livingstone D. M. , Hampton J. L. , Phipps P. M. , and Grabau E. A. 2005 Enhancing resistance to Sclerotinia minor in peanut by expressing a barley oxalate oxidase gene. Plant Physiol 137 : 1354 – 1362 .
Magbanua Z. V. , Wilde H. D. , Roberts J. K. , Chowdhury K. , Abad J. , Moyer J. M. , Wetzstein H. Y. , and Parrott W. A. 2000 Field resistance to tomato spotted wilt virus in transgenic peanut (Arachis hypogaea L.) expressing an antisense nucleocapsid gene sequence. Mol. Breed 6 : 227 – 236 .
McKently A. H. 1991 Direct somatic embryogenesis from axes of mature peanut embryos. In Vitro Cell. Dev. Biol. Plant 27 : 197 – 200 .
Melouk H. A. and Shokes F. M. 1995 Peanut health management APS Press St. Paul, MN .
Meyer P. 2000 Transcriptional transgene silencing and chromatin component. Plant Mol. Biol 43 : 221 – 234 .
Murray M. G. and Thompson W. F. 1980 Rapid isolation of high-molecular-weight plant DNA. Nucleic Acids Res 8 : 4321 – 4325 .
Ozias-Akins P. , Schnall J. A. , Anderson W. F. , Singsit C. , Clemente T. E. , Adang M. J. , and Weissinger A. K. 1993 Regeneration of transgenic peanut plants from stably transformed embryogenic callus. Plant Sci 93 : 185 – 194 .
Picard M. , Gross J. , Lubbert E. , Tolzer S. , Krauss S. , van Pée K-H. , and Berkessel A. 1997 Metal-free bacterial haloperoxidases as unusual hydrolases: activation of H2O2 by the formation of peracetic acid. Angew. Chem. Int. Ed. Engl 36 : 1196 – 1199 .
Rajasekaran K. , Cary J. W. , Jaynes J. M. , and Cleveland T. E. 2005 Disease resistance conferred by the expression of a gene encoding a synthetic peptide in transgenic cotton (Gossypium hirsutum L.) plants. Plant Biotechnol. J 3 : 545 – 54 .
Rajasekaran K. , Cary J. W. , Cotty P. J. , and Cleveland T. E. 2008 Development of a GFP-expressing Aspergillus flavus strain to study fungal invasion, colonization, and resistance in cottonseed. Mycopathologia 165 : 89 – 97 .
Rajasekaran K. , Cary J. W. , Jacks T. J. , Stromberg K. , and Cleveland T. E. 2000 Inhibition of fungal growth in planta and in vitro by transgenic tobacco expressing a bacterial nonheme chloroperoxidase gene. Plant Cell Rep 19 : 333 – 338 .
Rohini V. K. and Rao K. S. 2001 Transformation of peanut (Arachis hypogaea L.) with tobacco chitinase gene: variable response of transformants to leaf spot disease. Plant Sci 160 : 889 – 898 .
Sambrook J. , Fritsch E. F. , and Maniatis T. 1989 Molecular cloning, a laboratory manual Cold Spring Harbor Laboratory Press Cold Spring Harbor, NY .
Samoylov V. M. , Tucker D. M. , and Parrott W. A. 1998 Soybean [Glycine max (L.) Merrill] embryogenic cultures: the role of sucrose and total nitrogen content on proliferation. In Vitro Cell. Dev. Biol. Plant 34 : 8 – 13 .
Sharma K. K. and Anjaiah V. 2000 An efficient method for the production of transgenic plants of peanut (Arachis hypogaea L.) through Agrobacterium tumefaciens-mediated genetic transformation. Plant Sci 159 : 7 – 19 .
Singsit C. , Adang M. J. , Lynch R. E. , Anderson W. F. , Wang A. , Cardineau G. , and Ozias-Akins P. 1997 Expression of a Bacillus thuringiensis cryIA(c) gene in transgenic peanut plants and its efficacy against lesser cornstalk borer. Transgenic Res 6 : 169 – 176 .
Topfer R. , Maas C. , Horicke-Grandpierre C. , Schell J. , and Steinbiss H-H. 1993 Expression vectors for high-level gene expression in dicotyledonous and monocotyledonous plants. Meth. Enzymology 217 : 66 – 78 .
van Pée K-H. 1996 Biosynthesis of halogenated metabolites by bacteria. Ann. Rev. Microbiol 50 : 375 – 399 .
Wang A. , Fan H. , Singsit C. , and Ozias-Akins P. 1998 Transformation of peanut with a soybean vspB promoter-uidA chimeric gene. I. Optimization of a transformation system and analysis of GUS expression in primary transgenic tissues and plants. Physiol. Plant 102 : 38 – 48 .
Wiesner W. , van Pée K-H. , and Lingens F. 1988 Purification and characterization of a novel bacterial nonheme chloroperoxidase from Pseudomonas pyrrocinia. J. Biol. Chem 263 : 13725 – 13732 .
Wilson D. M.
1995
Management of mycotoxins in peanut.
87
–
92
In
Melouk H. A.
and
Shokes F. M.
Peanut Health Management
APS Press
St Paul, MN
.
Wolfframm C. , Lingens F. , Mutzel R. , and van Pée K-H. 1993 Chloroperoxidase-encoding gene from Pseudomonas pyrrocinia: sequence expression in heterologous hosts, and purification of the enzyme. Gene 130 : 131 – 135 .
Yang H. , Singsit C. , Wang A. , Gonsalves D. , and Ozias-Akins P. 1998 Transgenic peanut plants containing a nucleocapsid protein gene of tomato spotted wilt virus show divergent levels of gene expression. Plant Cell Rep 17 : 693 – 699 .
Yang H. , Ozias-Akins P. , Culbreath A. K. , Gorbert D. W. , Weeks J. , Mandal B. , and Pappu H. 2004 Field evaluation of tomato spotted wilt virus resistance in transgenic peanut (Arachis hypogaea, L.). Plant Dis 88 : 259 – 264 .
Notes
Author Affiliations
1Department of Horticulture, The University of Georgia Tifton Campus, Tifton, GA 31793-0748, USA.
2Food and Feed Safety Research Unit, USDA, ARS, Southern Regional Research Center, New Orleans, LA 70124.
3Present address: Department of Plant and Soil Science, Texas Tech University, Lubbock, TX 79409, USA.
4Present address: Department of Agriculture, Iwate University, Morioka, Iwate 020-8550, Japan.
5Present address: Faculty of Agricultural Sciences, Catholic University of Córdoba, Camino a Alta Gracia km 7 1/2 (5017) Córdoba, Argentina.
6Present address: Department of Plant Pathology, Cornell University, Ithaca, NY 14853, USA.
7Present address: College of Veterinary Medicine, University of Georgia, Athens, GA 30602, USA.
*Corresponding author: pozias@uga.eduHygromycin phosphotransferase
CaMVCauliflower mosaic virus
CPO-PChloroperoxidase from Pseudomonas pyrrocinia